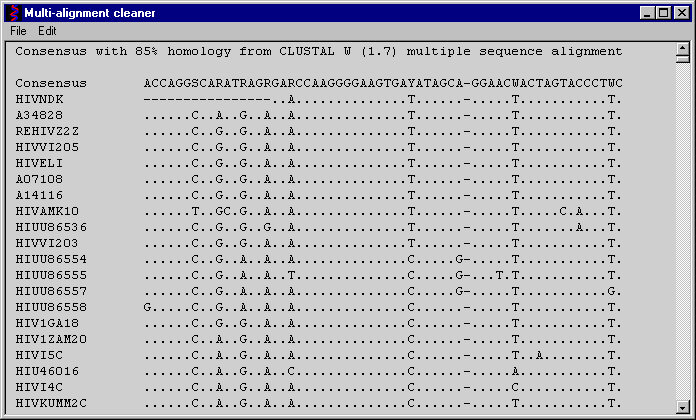
Check on the "Select a reference sequence" radio button
Choose the sequence do you want to use as a reference for the multi-alignment in the list box
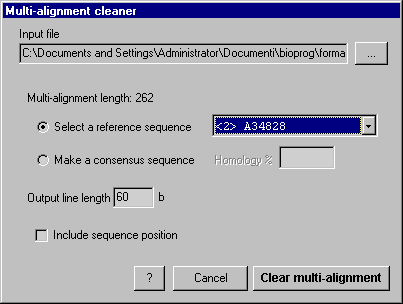
Check the "Make a consensus sequence" radio button
Enter the homology percent for the consensus
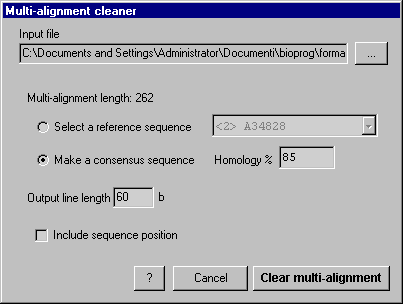
| AnnHyb's help - Multi-alignment cleaner |