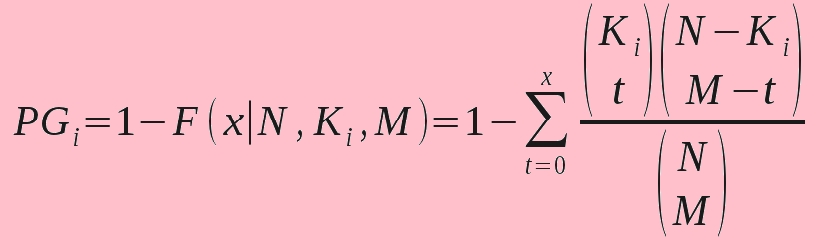The microRNA-mRNA file should contain microRNA and mRNA pair in each line separated by \t (Tab) which starts with microRNA. Similarly mRNA-GO file should contain mRNA and GO pair in each line separated by \t (Tab) which starts with mRNA.
Steps to create microRNA-mRNA file
- Go to PMRD database
- Click on 'download' then right click on your plant microRNA's 'mature sequence' to choose 'Save Target As...' . Suppose the file name be xxx_mature.txt
- Go to psRNATarget server and click on 'User-submitted small RNAs / preloaded transcripts' (this is already selected as default)
- Click on 'Choose File' and then browse xxx_mature.txt
- Then from the given options in 'select a preloaded transcript/genomic library for target search' input box select corresponding option.
- Click on 'Submit' with default parameters or values as required.
- Click on 'Batch Download' and Save the file.
- Open the saved file in excel and delete all columns except the first two then save the file in csv/tsv text file
Steps to create mRNA-GO file
- Go to Biomart EnsemblPlants page.
- Choose 'Ensembl Plants Genes nn' from CHOOSE DATABASE drop-down box.
- Choose dataset of your plant from CHOOSE DATASET drop-down box.
- Click on the Attributes on the left hand panel
- Expand GENE by clicking on the + sign beside GENE: in the right panel
- Deselect 'Ensembl Gene ID'
- Expand External and select 'GO Term Accession (bp)'
- Click on 'Results' button
- Select the 'Unique results only' checkbox and click on 'Go' and save the file
Note: mRNA ids present in microRNA-mRNA and mRNA-GO files should be of same id category, e.g., both mRNA id from Refseq or Ensembl or NCBI etc.
Steps to use the software
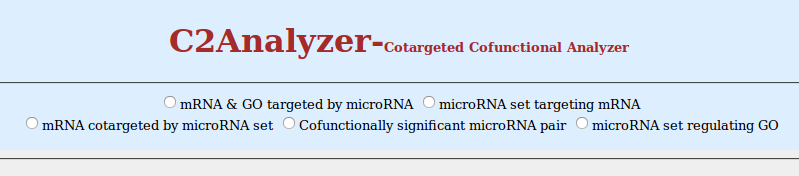
1. mRNA & GO targeted by microRNA
- Click on radio button of 'mRNA & GO targeted by microRNA'
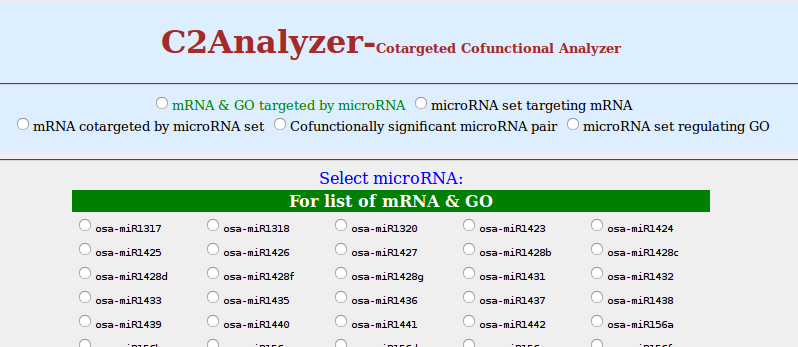
- Select microRNA of your interest by clicking on the radio button
Here first table shows microRNA you selected, total number of mRNA targeted by that microRNA and list of mRNA respective columns. Second table shows microRNA, the number of GO biological processes and list of GO ids. GO ids are corresponding to the target mRNA.
2. microRNA set targeting mRNA
- Click on radio button of 'microRNA set targeting mRNA'
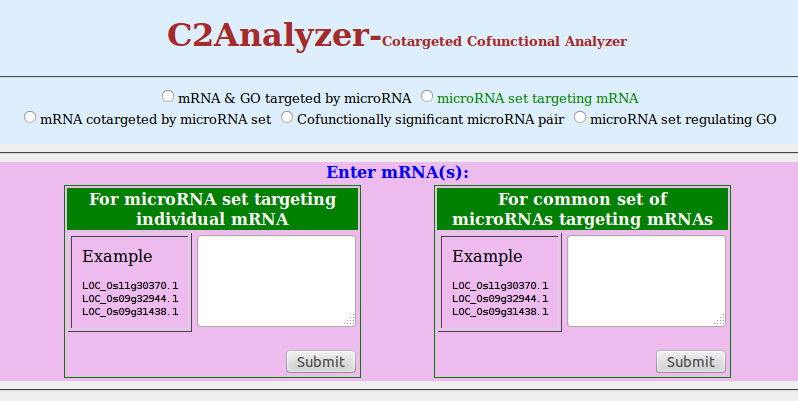
- You can enter mRNA in two different boxes as per requirement. If you want to determine a set of microRNAs which target mRNA(s), choose the left textbox. If you want to get a list of microRNAs cotargeting a set of given mRNAs, then choose the right textbox. You can enter mRNAs separated by comma or newline.
- Click on 'Submit'
Find your results on respective columns
3. mRNA cotargeted by microRNA set
- click on radio button of 'mRNA cotargeted by microRNA set'
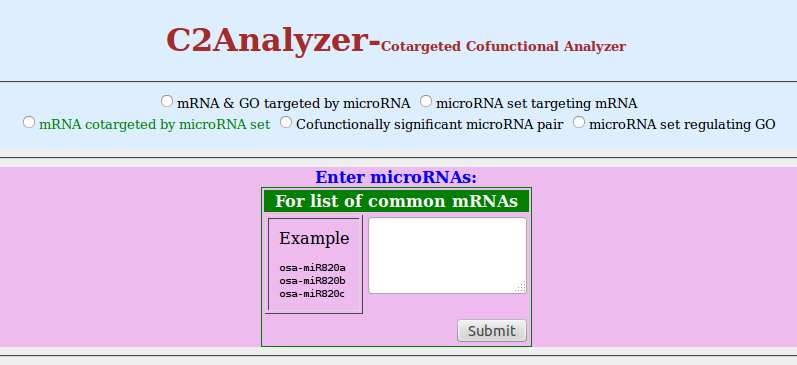
- You can enter microRNA set of your interest to find out the mRNA set cotargeted by them
- Click on 'Submit'
4. Cofunctionally significant microRNA pair
- Click on radio button of 'Cofunctionally significant microRNA pair'
Note: For a given microRNA pair of microRNA A and B, we identified the target subset they co-target (A ∩ B). The subset is required to have at least Omin genes. First, biological processes where the target subset is enriched are identified by hypergeometric distribution. The probability PGi for A ∩ B in the GO term i is calculated according to the formula (Xu J et al., 2011) given below
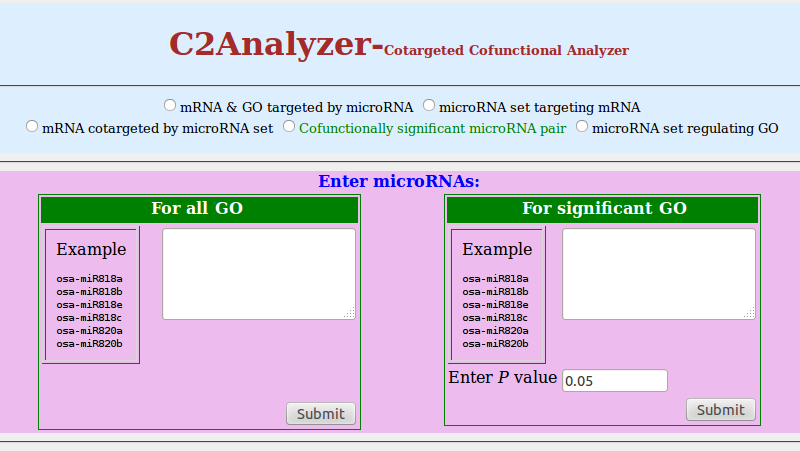
- You can enter microRNAs in two different boxes as per requirement. If you want to determine a set of GO(s) which are regulated by pairs of microRNAs, choose the left textbox. If you want to obtain the set of GO(s) with enriched targets regulated by pairs of microRNAs then choose the right textbox. You can enter microRNAs separated by comma or newline. For the second case, enter P-value of your choice in the box [default value is 0.05]
- Click on 'Submit'
5. microRNA set regulating GO
- Click on radio button of 'microRNA set regulating GO'
Note: To identify the microRNAs, whose targets are enriched in the given GO id(s), χ2 test has been performed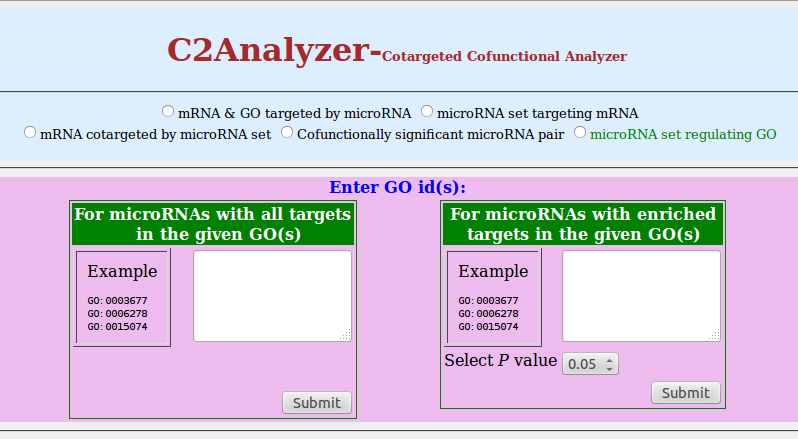
- You can enter GO ids in two different boxes as per your requirement. To determine a set of microRNAs which are involved in single GO process, enter GO id(s) in left textbox. To obtain the set of microRNAs which regulate your set of GO processes with enriched targets, enter GO id(s) in right textbox.
- Click on 'Submit'