Home
Documentation
FindtRNA
version 1.0Introduction:
In archaea and eukaryotes approximately 15% tRNA precursors contain intron(s). Although most of them are located at the unique canonical position 37 & 38(37/38), recent report indicates that introns are also located in various non-canonical positions. TRNA-scan-SE and ARAGORN are widely used that identify tRNA genes without introns or with introns at canonical positions. FindtRNA able to identifies tRNA genes without introns or with introns at canonical positions or also at non-canonical positions.The program employs algorithm to find number of tRNA present in the supplied genome sequence and predict the tRNA secondary (cloverleaf) structure of those tRNAs.
Download:
Download the software from here
Tutorial:
Panels::
Input panel
In the input panel there is a TextArea where user can paste the fasta format of the sequence. Or a file, saved in fasta format, can be loaded by the browse button.
After pasting the sequence or attaching the file, submit button have to be pressed. Whenever the calculation would be over, the result panel and the image panel would be available.
Result panel
By clicking the result tab on the top one can go to the result panel to know how many tRNAs have been found.
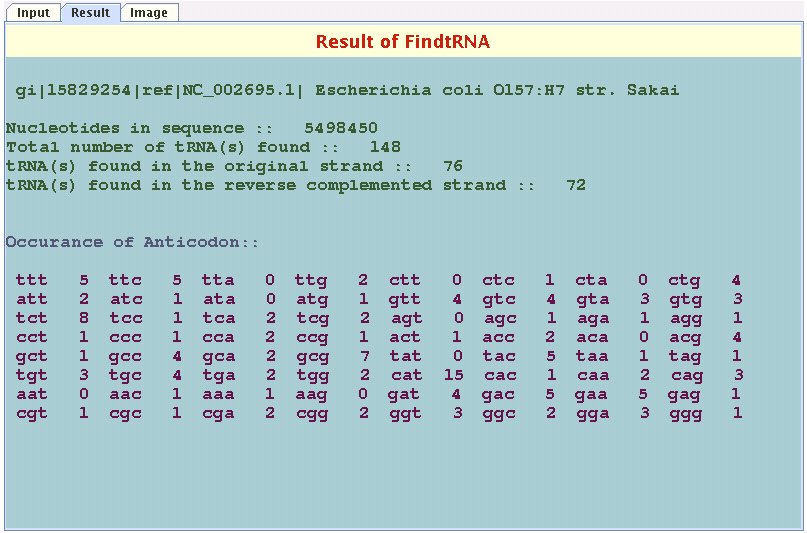
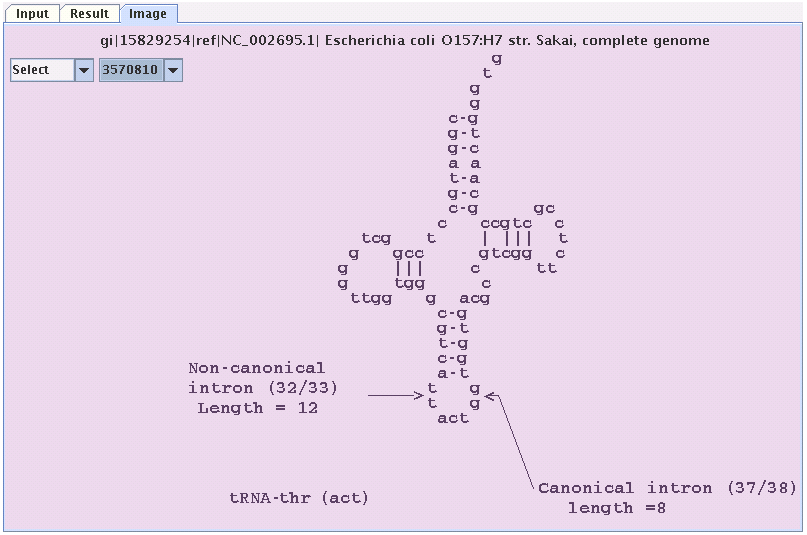
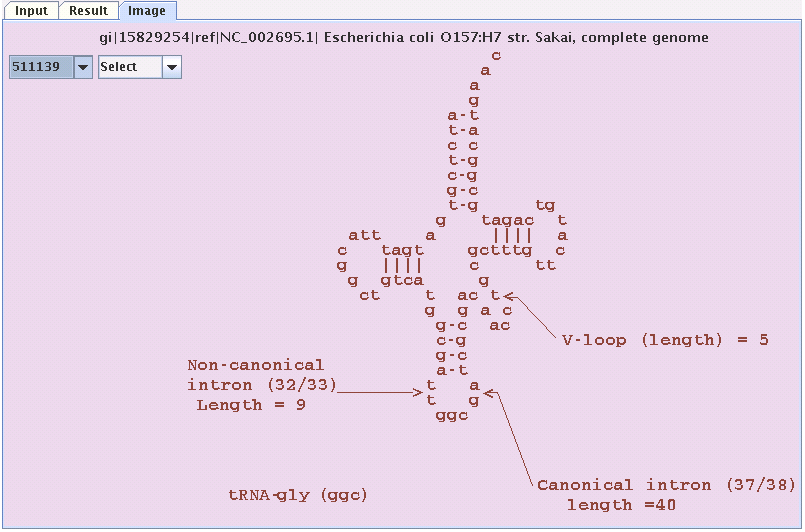
Image panel
Similarly by clicking the image tab one can see the coverleaf structure of
the tRNA(s) that have been found by FindtRNA.In this panel there are two combo boxes
one for the original strand and one for the reversed complemented strand.by clicking
the positions in the combo boxes one can see the structure of the tRNA at that position
on the sequence.
Selecting a position in the original strand
Selecting a position in the reverse complemented strand