- Click the radio button to select Inter or Intra Kingdom
- Select miRNA and Target from the drop down menu
- Set the E-cutoff and click 'Submit'
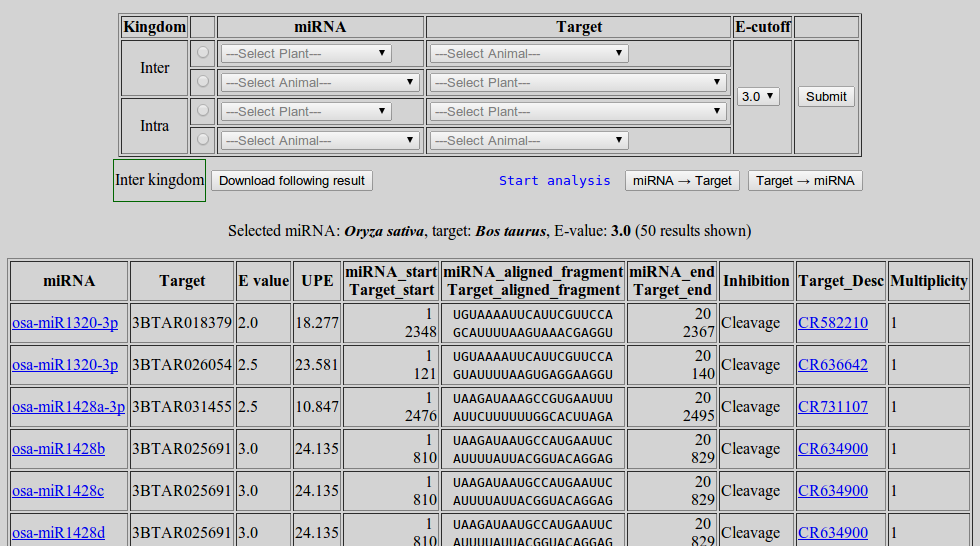
The results will be displayed. At most 50 results will be shown on the screen. However the full set of results can be saved. One can navigate to the description of miRNA(s) and target(s) by clicking the respective link(s)
- To save the full result click 'Download following result'.
- Next two types of analysis can be performed:
- miRNA → Target
- Target → miRNA
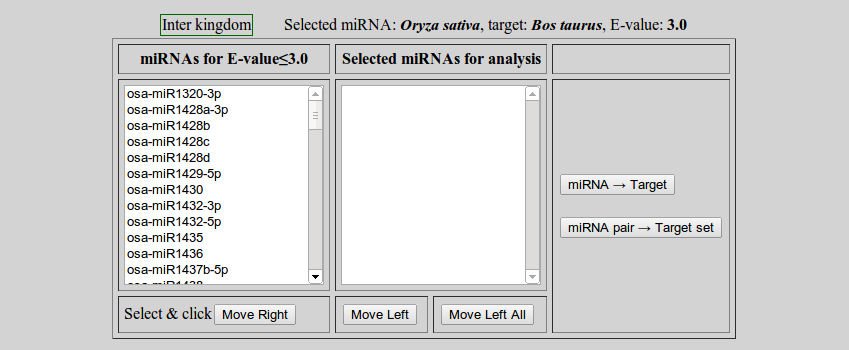
To start miRNA to target analysis click 'miRNA → Target'. A appearence will change. The left box contains the list of miRNAs at user defined expectation value.
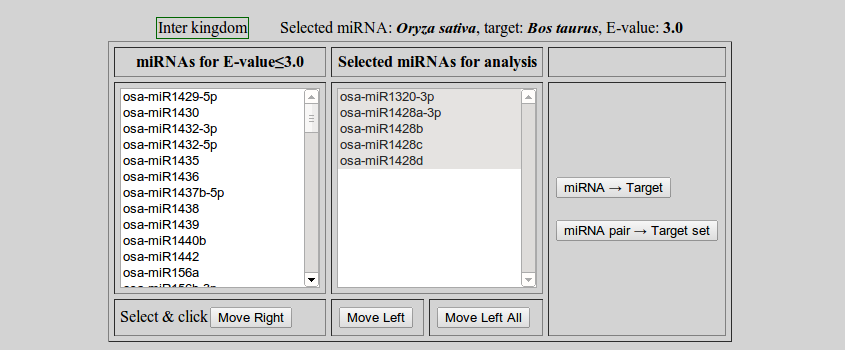
To analyze the miRNA(s) of interest the user must move the miRNA(s) from left box to'Selected miRNAs for analysis' box.
- Select miRNA(s) of interest and click 'Move Right'. The user may use CTRL+Click or SHIFT+Click for multiple selection
- To remove miRNA(s) from 'Selected miRNAs for analysis' box, select the miRNA(s) and click 'Move Left'
- Here two types of analysis can be done
- miRNA -> Target
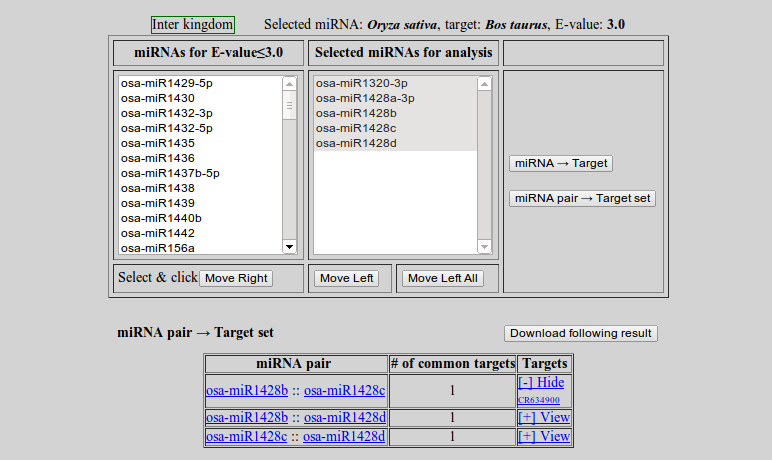
- miRNA pair -> Target set
To start miRNA to target analysis click 'miRNA → Target'. The results will appear below.
- To save the results click 'Download following result'
- To view the targets click '[+] View'
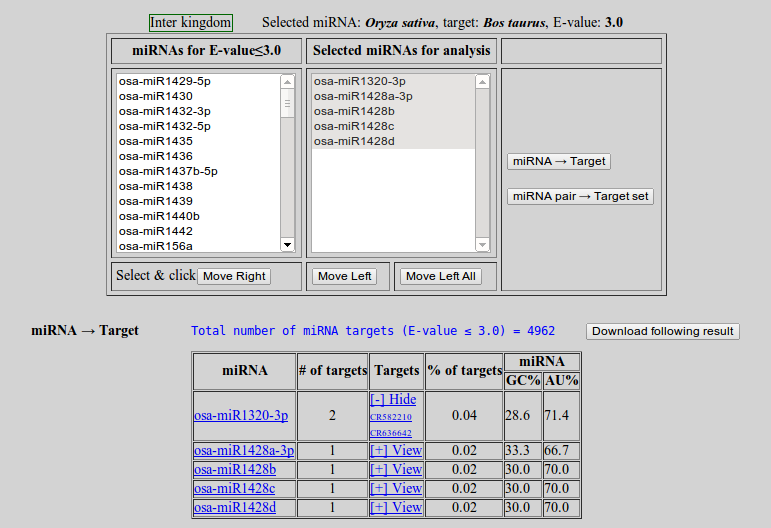
To perform the other type of analysis click 'miRNA pair → Target set'
- To save the result click 'Download following result'
- To view the targets click '[+] View'
Similarly Target to miRNA analysis can be performed by clicking 'Target → miRNA'.