Teachers:
Here
is a 2016 syllabus for an 8-hour block (3 afternoons in a computer lab) in a
lab course for senior undergraduates. Each student
picks a molecule from this Atlas, then reports on how its structure supports its function.
The report has eleven sections (questions) and a sample completed report is provided.
Feel free to copy/adapt:
Proteopedia encourages
share-alike re-use with attribution, as does this Atlas (see
license at top).
Proteopedia offers an article on
Teaching Strategies that includes suggested
lesson plans.
In each category below, PDB files have been divided into
those that are relatively straightforward, those that
are more challenging, and sometimes enormous.
"Straightforward"
cases have usually been selected to avoid complications (such as being
very large,
lacking sidechains,
having many
alternative sidechain conformations, etc.).
Below, Years in parentheses after links to molecules
indicate the years of publication. In some cases a range of years
is given: the early year is when the molecule was first solved
(if I happen to know that)
at 3.5 Å
resolution
or better;
the later year is when the structure chosen for the Atlas was published.
Update History/Versions.
|









 CHALLENGING
CHALLENGING




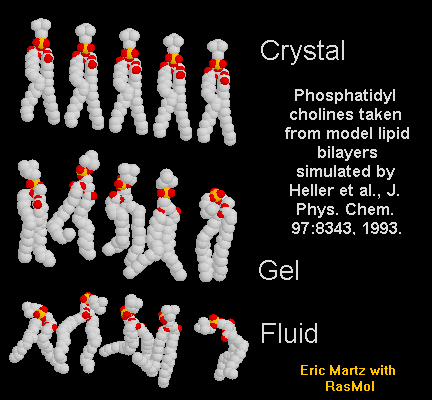 (Yes, we know they're not really macromolecules.)
(Yes, we know they're not really macromolecules.)


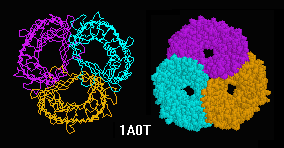



 Transfer RNA (Phe),
(
Transfer RNA (Phe),
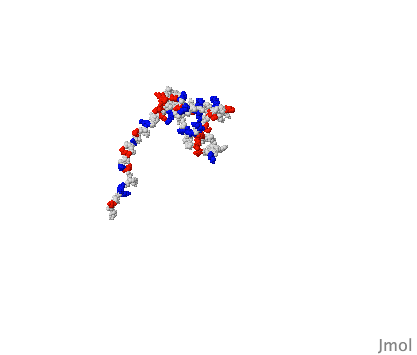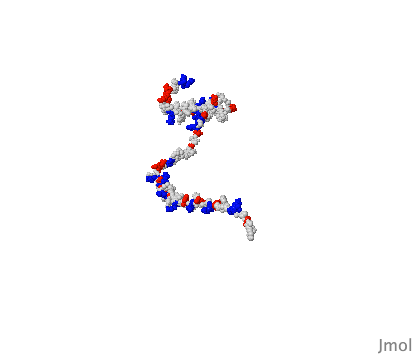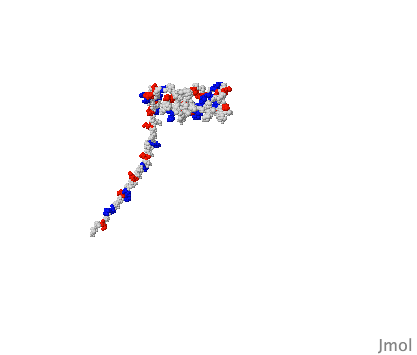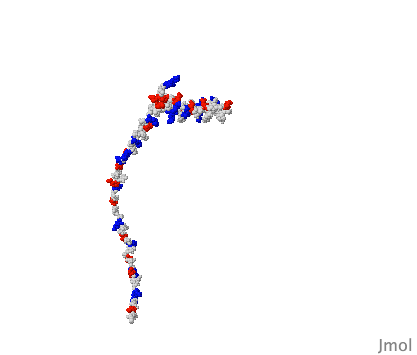
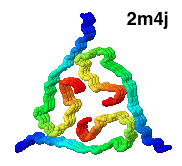
(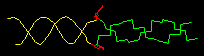 DNA B form to Z form transition,
(2005).
The cover of the issue of Nature reporting this structure
showed a theoretical model with longer B- and Z-form ends (shown at right),
kindly provided for this Atlas by Kyeong Kyu Kim.
View this model
In order to solve this structure, the Z-DNA portion was stabilized
by Z-DNA-binding proteins.
The surprise is that base pairing is disrupted at only a single pair at
the transition point.
Sequences near promotors often favor Z-DNA, enabling them to trap
negative supercoiling that occurs behind a moving polymerase, or
during nucleosome unwrapping. Z-DNA cannot participate in a nucleosome,
hence exposing it to transcription factors.
DNA B form to Z form transition,
(2005).
The cover of the issue of Nature reporting this structure
showed a theoretical model with longer B- and Z-form ends (shown at right),
kindly provided for this Atlas by Kyeong Kyu Kim.
View this model
In order to solve this structure, the Z-DNA portion was stabilized
by Z-DNA-binding proteins.
The surprise is that base pairing is disrupted at only a single pair at
the transition point.
Sequences near promotors often favor Z-DNA, enabling them to trap
negative supercoiling that occurs behind a moving polymerase, or
during nucleosome unwrapping. Z-DNA cannot participate in a nucleosome,
hence exposing it to transcription factors.
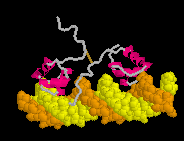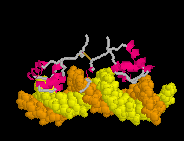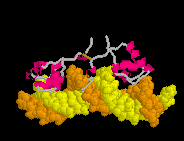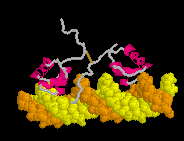




 Defensin (Rhesus theta defensin one, RTD-1), an antibacterial
cyclic 18-amino acid peptide,
(20-model NMR structure, see
Defensin (Rhesus theta defensin one, RTD-1), an antibacterial
cyclic 18-amino acid peptide,
(20-model NMR structure, see  Since these models contain no protein and no nucleic acid chains, the entire model is
Ligands+ in FirstGlance. The Vines/Sticks view is useful. Toggle the Ligands+
button to spacefill.
Since these models contain no protein and no nucleic acid chains, the entire model is
Ligands+ in FirstGlance. The Vines/Sticks view is useful. Toggle the Ligands+
button to spacefill.