Discover OligoFaktory Standalone Edition

The user interface emphasizes useability and is aimed at assisting researchers for a painless, rapid, automated, and reliable design.
Three different view modes
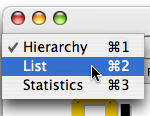
You will be able to visualize your results in three different flavours:
- The Hierarchy view is a long listing that shows relative location bargraphs of queries and results.
- The List view is a short listing that emphasizes leaf results, while hiding locations information.
- The Statistics view summarizes the results with charts showing the distributions of main features.
Emphasize important features

The output includes the list of oligo sequences together with their corresponding locations on the query sequences, their lengths, and their melting temperatures.
Easy-to-spot warning flags are shown in case of problems with secondary structures and/or with specificity evaluation.
Details available in one click
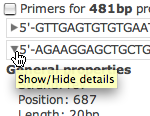
All detailed information on heterodimer, homodimer, hairpin, sensitivity, and specificity unfold by clicking on the triangle in front of the oligonucleotide sequence.
Search your results
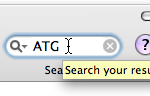
The search field highlights any occurence of a given string in the result view.
Encoder for query sequences and regions
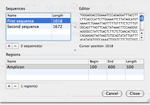
A master-detail interface allow users to enter their sequences and related regions.
Users can cut-and-paste query DNA into the Editor view, or they can use the File > Import… menu item to import FASTA files.
Five complementary actions
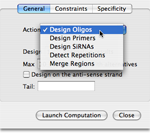
Several design iterations can be applied, using five complementary actions:
- The Design Oligos action allows for the automated design of specific long oligonucleotides on an arbitrary number of ORFs (e.g., for the development of DNA microarrays).
- The Design Primers action allows for the automated design of specific PCR primer pairs on an arbitrary number of regions.
- The Design SiRNAs action allows designing optimal 19bp siRNA with the method described in Rational siRNA design for RNA interference, A. Reynolds et al. Nature biotechnology, 2004, vol 22, 326-330.
- The Detect Repetitions action identifies all repetitions and microsatelites under user-specified constraints.
- The Merge Regions action merges regions which are too close to allow designing specific primer pairs.
Design constraints
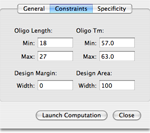
For design actions, the user has the possibility to control various constraints:
- The accepted range of oligonucleotide length.
- The accepted range of product length for PCR.
- The accepted range of melting temperatures.
- The preferred location of the oligonucleotides.
- The presence of a 5'-end tail.
Specificity evaluation
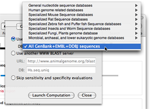
If needed, you can choose a species against which oligonucleotide sensitivity and specificity will be evaluated.
When choosing that option, the software will check whether any oligonucleotide exhibits a significant match with other target in the database.
You can choose a database hosted on the NCBI server. You can also use your own WWW BLAST server and your custom databases.
Export to various file formats
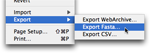
You can export results to FASTA or a comma delimited file (CSV), readable by most spreadsheet programs such as Microsoft Excel. You can also export the current view to a web archive compatible with Safari.
Advanced controls
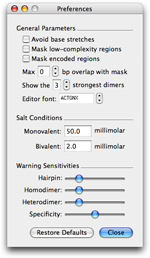
Shared parameters and advanced controls are availables in the preferences panel:
- The General Parameters options let you define masking policies among other things.
- The Salt Conditions can be tweaked to calibrate the melting temperature (Tm) estimation.
- The Warning Sensitivities options let you define the sensitivity of warning flags raised in the listings.

This free software was created by Colas Schretter, 2005-2009.
Feel free to contact him at oligofaktory [at] gmail [dot] com