Design primers with OligoFaktory Standalone Edition

Learn with a simple tutorial for designing primer pairs for PCR amplifications. Click on thumbnails to view full size images.
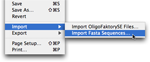
Import FASTA sequences
Click the File Import menu item to import FASTA files containing query sequences.
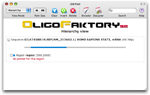
Many query sequences can be processed in batch. However, for this tutorial we will use the following DNA sequence.
Encode a region
Click on the Encoder icon of the toolbar and add a region for the selected sequence.
Specify the location and the length for the region. For example, set begin at base 300, and a length of 1700bp.
View sequences and regions
After closing the encoder, a range corresponding to the region is highlighted in blue.
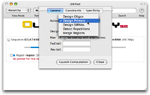
Select a design tool
Click on the Designer icon of the toolbar and select the Design Primers action.
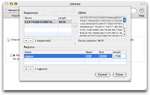
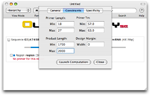
Specify design constraints
Select the Constraints tab and specify the accepted range for product length.
Since the length of the region to amplify is 1700bp, the product length is chosen to lie between 1700bp and 2000bp. The other parameters are left untouched.
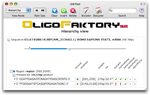
Launch computation
Click on the Launch Computation button. Et voila, a pair of primers is produced. If nothing appears, this probably mean that the design constraints are too stringent.
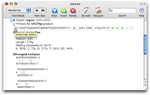
View detailed information
Click on a triangle in front of any oligonucleotide sequence and detailed information will unfold below.

This free software was created by Colas Schretter, 2005-2009.
Feel free to contact him at oligofaktory [at] gmail [dot] com