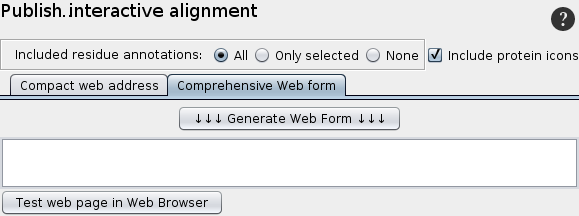
Publish interactive alignment
With this dialog a Web-Link can be formed which loads the proteins from the public databases into Strap and displays an alignment.
The only requirement for the client machine is Java.
Depending on the amount of information two different types are
available:
1. Single web address
The web address can not only be included in web-pages, but also in e-mails and Office-documents.
The generation is conducted in two steps:
-
 The
parameter String is written into the first text field and can
be modified by the user. Not the sequence but the database
accession id is stored. For technical details see
http://www.bioinformatics.org/strap/createStrapLinks.html.
The
parameter String is written into the first text field and can
be modified by the user. Not the sequence but the database
accession id is stored. For technical details see
http://www.bioinformatics.org/strap/createStrapLinks.html.
-
 From the
text in the first text field the web address will be generated
using and written into the 2nd text field.
From the
text in the first text field the web address will be generated
using and written into the 2nd text field.
 The
generated web address can be tested by clicking. A new STRAP
session will be opened in web-mode and an alignment will be
loaded with the specified information.
The
generated web address can be tested by clicking. A new STRAP
session will be opened in web-mode and an alignment will be
loaded with the specified information.
2. Web form
Since the web form has no size limitation, the entire information for the alignment can be included.
The draw-back is that it can only be included in web-pages, but not in office documents or e-mails.
 A html-page including the web link for the selected proteins is generated.
A html-page including the web link for the selected proteins is generated. The link can be tested in the web browser.
From this html code the text between the opening and closing and <body> tags can be used in any html-page.
The link can be tested in the web browser.
From this html code the text between the opening and closing and <body> tags can be used in any html-page.

 The
parameter String is written into the first text field and can
be modified by the user. Not the sequence but the database
accession id is stored. For technical details see
http://www.bioinformatics.org/strap/createStrapLinks.html.
The
parameter String is written into the first text field and can
be modified by the user. Not the sequence but the database
accession id is stored. For technical details see
http://www.bioinformatics.org/strap/createStrapLinks.html.
 From the
text in the first text field the web address will be generated
using and written into the 2nd text field.
From the
text in the first text field the web address will be generated
using and written into the 2nd text field.
 The
generated web address can be tested by clicking. A new STRAP
session will be opened in web-mode and an alignment will be
loaded with the specified information.
The
generated web address can be tested by clicking. A new STRAP
session will be opened in web-mode and an alignment will be
loaded with the specified information.  A html-page including the web link for the selected proteins is generated.
A html-page including the web link for the selected proteins is generated. The link can be tested in the web browser.
From this html code the text between the opening and closing and <body> tags can be used in any html-page.
The link can be tested in the web browser.
From this html code the text between the opening and closing and <body> tags can be used in any html-page.