 .
Those entries usually contain the variables "PROTEIN" and "RESIDUES"
which stand for the current protein number and the sequence positions.
.
Those entries usually contain the variables "PROTEIN" and "RESIDUES"
which stand for the current protein number and the sequence positions.
\feature{top}{PROTEIN}{RESIDUES}{box[Red]}{peptide}
\feature{ttop}{PROTEIN}{RESIDUES}{box[Red]}{peptide}
\feature{bottom}{PROTEIN}{RESIDUES}{fill:$\uparrow$}{$S_{129}$}
\feature{bbottom}{PROTEIN}{RESIDUES}{fill:X}{cleavage}
\fingerprint{300}
|
\feature{bottom}{PROTEIN}{RESIDUES}{fill:$\uparrow$}{$$FIRST_Aaa_{$FIRST_INDEX}$}
\feature{bottom}{PROTEIN}{RESIDUES}{fill:$\uparrow$}{$$FIRST_A_{$FIRST_INDEX}$}
|
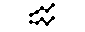 .
The plot has four alternative locations: top, bottom, ttop, bbottom.
.
The plot has four alternative locations: top, bottom, ttop, bbottom.
# fmtutil --byfmt=pdflatex