Tutorials
How to install Assemble2?
Assemble2 is divided into two parts:
- a graphical tool (which is the only part that need to be installed for "classical" users),
- RNA algorithms and RNA data. For several tasks (2D prediction, 3D annotation,...), Assemble2 delegates the work to a bunch of Web services. By default, Assemble2 is configured to use our public server (http://arn-ibmc.in2p3.fr/api/). But you can also deploy your own server locally to increase the speed of your Assemble2 and of all the Assemble2 programs running on your local network.
In this tutorial, you will learn how to:
Installation of the graphical tool (mandatory)
To use Assemble2, you will need:
- a Java environment installed on your computer. You need to have Java version 8 update 20 (8u20) or greater (non-official releases of Java are not supported (gcj, cacao, kaffe,...)). Open a terminal or a console and type:
java -version
If this command is not found by your system, or if you don't have Java version 8 update 20 (8u20) or greater, you need to download and install it. Simply go to the Java website. The last one is the best ;-) .
- an internet connection (Assemble2 uses several Web services),
- a 3-buttons mouse,
- the software UCSF Chimera installed.
First, download the last stable or development release. Once the archive unzipped, double-click on the file launch_Assemble2_for_XXX or launch it from a terminal.
After the first launch of Assemble2, a window will popup, asking you to set up the path of the Chimera executable. Once done, Assemble2 will be displayed, along with a Chimera window. If Chimera is not launched automatically, you can go to "File -> Configure... -> Assemble2" to fix the path of Chimera (see here for details).
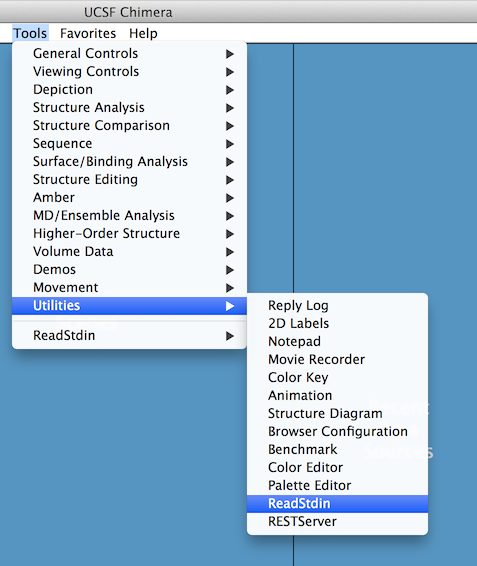
Next, you need to open the "communication channel" of Chimera by selecting "Tools -> Utilities -> ReadStdin".
Once done, go to "File -> Load... -> RNA Tertiary Structure... -> From a PDB file", and choose the file 1ehz.pdb in the directory "samples/tertiary structures". If everything goes well, you should have something like that:

To greatly improve your efficiency in RNA 3D modeling, we suggest you to plug a device like the Space Navigator from 3DConnexion. Use the Space Navigator to move the entire 3D model, and configure Chimera to use the mouse to move the selection only (Tools -> Movement -> Movement Mouse Mode).
Deploy your own Web services (optional)
You need to deploy an RNA Science Toolbox on your computer. You will find all the details on this page.

