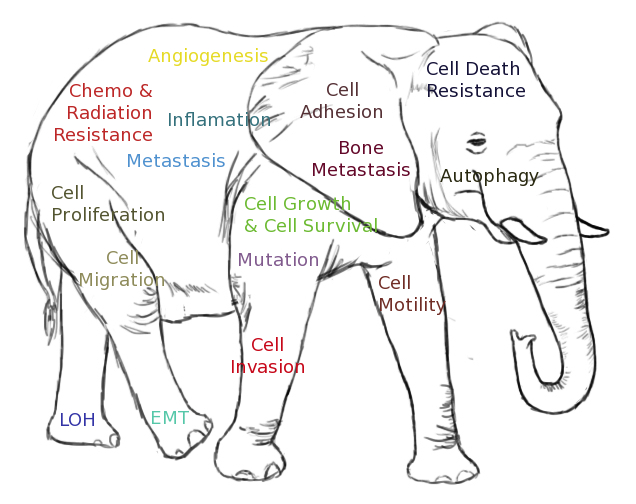
Prostrate Cancer Hallmarks Map is an initiative taken from a system’s medicine perspective for detail systems level functional exploration of human prostate cancer specifically at molecular level with literature mined knowledge and insights. The principal aim of this project lies in the reconstruction of signalling pathway maps & altered circuitries of various cell biological events associated with the pathogenesis of human prostate cancer through a network medicine based approach. It represents signalling pathway based synthetic pictures of different prostate cancer hallmarks and their underlying oncopathological and cell biological features.
Prostate cancer is one of the major leading causes of cancer related mortality in male population of the Western Europe and United States. Patients with metastatic castration resistant prostate cancer are expected to survive not more than nineteen months. This is due to the facts that there is no significantly effective molecularly targeted therapy available for prostate cancer. Androgen deprivation therapy remains the only treatment strategy for locally advanced to metastatic prostate cancer, but majority of the patients will eventually progress into castration resistant disease form which is currently totally untreatable. It is assumed that androgen independent molecular pathways function as the key driver of castration resistant prostate cancer progression. Here we represent various androgen independent oncopathways associated with prostate cancer tumorigenesis and progression.
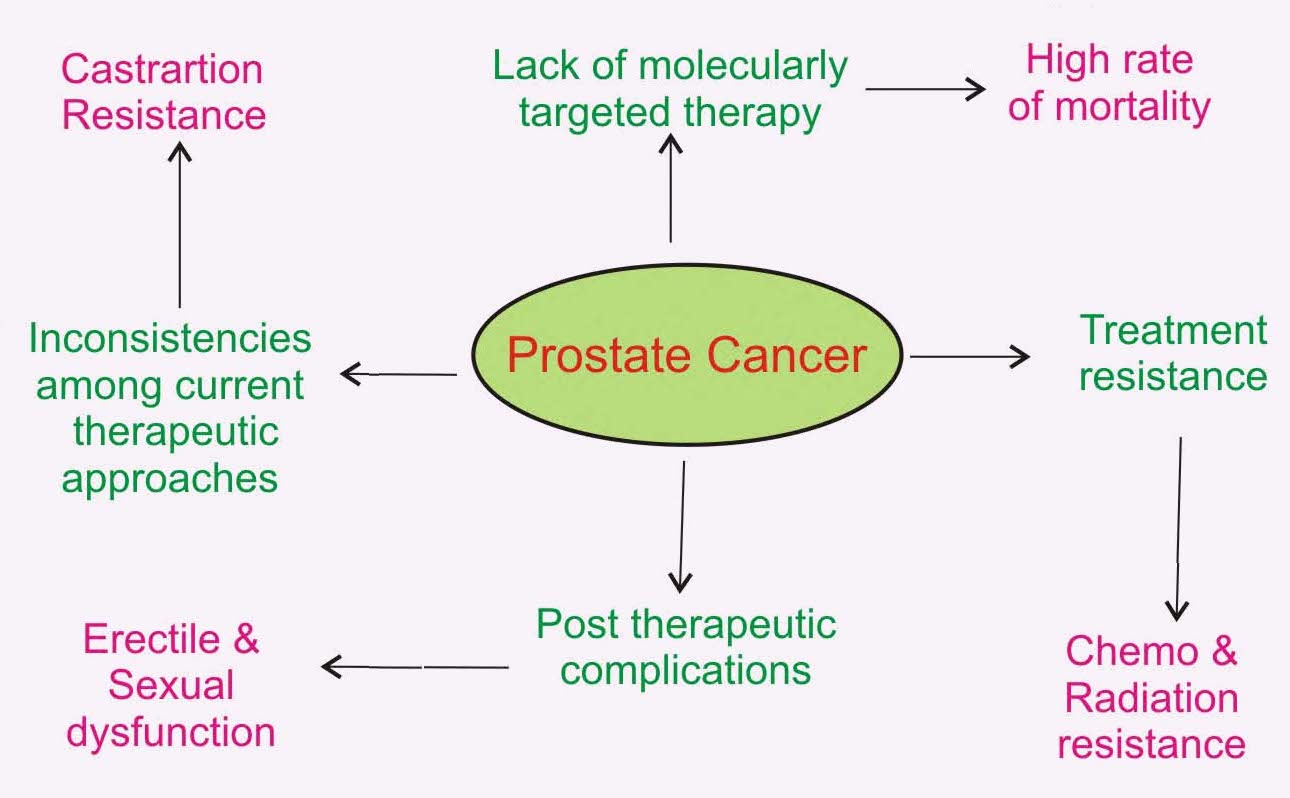
Prostate cancer at a glance- represents some basic facts around current prostate cancer treatment approach, its limitations and its associated post therapeutic complications.
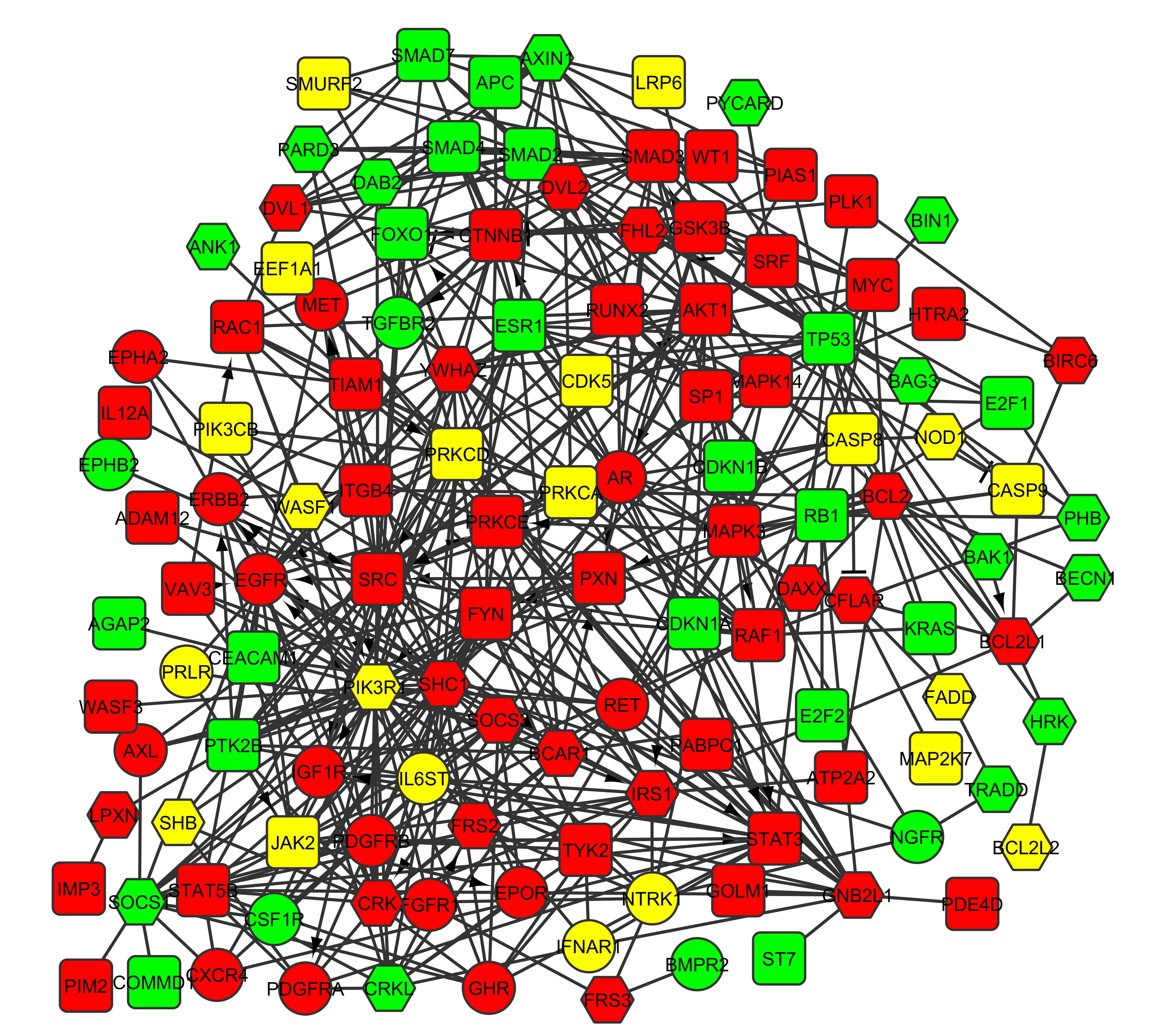
The journey in Prostrate Cancer Hallmarks Map has been started with the reconstruction of a molecular interactome network for human prostate cancer centered on adaptor protein. This adaptor centric prostate cancer associated molecular interactome has been reconstructed from biomedical information derived from published experimental studies through a manual literature curation based method. Our intent is to provide the basic cancer research community with a comprehensive resource for exploring molecular architecture of human prostate cancer at four specific levels, namely as information related to prostate cancer hallmark & its underlying feature based phenomenon, prostate cancer hallmark based pathway map, hallmark based events related functional molecular connectivity map and its underlying pathway based modular organizational map. Additionally, it also provides information related to prostate cancer inter hallmark based modular organization.
The core network of adaptor centric molecular interactome associated with human prostate cancer.