back to STRAP: multiple sequence alignment
also see Pymol
3D-superposition of proteins
Dialog protein structure Viewer and
Protein 3D-superposition with STRAP
algorithms to superimpose protein 3D structures are applied to identify similarities of
protein folds.
The coordinates of a mobile protein is transformed (superposed) so that the
backbone lies over the backbone of a reference protein.
Distant homologues may not be recognized by their amino acid sequence
because the sequences diverge more rapidly in evolution than the
3D-structure.
In these cases three-dimensional protein superposition of protein structures is very valuable.
Currently there are two procedures for protein superposition available in STRAP:
- Combinatorial Extension (CE) determines an optimal structure alignment between aligned fragment pairs (AFPs). AFPs are determined from local geometry averaged over 8 C alpha positions. Heuristics are used to prevent a combinatorial explosion.Final alignments are by dynamic programming.
- DIP-OVL written by Andrean Goede can superpose two sets of point in three-dimensional space. The algorithm does not need a specific order of the points. The connectivity of the protein backbone is neglected. The algorithm assigns points from one set to points to the other set minimizing the Euclidean distances (RMSD) of the assigned pairs.
In the form a mobile and a reference protein are
selected. Alternatively a hole bunch of mobile proteins can be
probed against one reference protein.

After superposition the protein backbones can be inspected three-dimensionally.

Pymol superposition: Per click superimposition of proteins can be displayed in Pymol
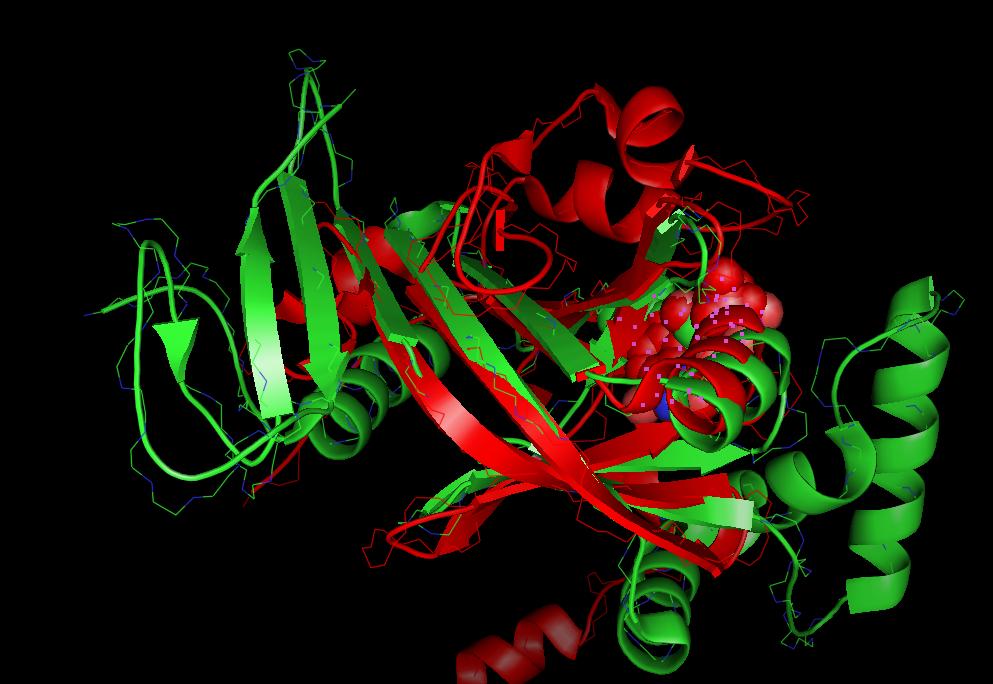
STRAP also provides a multiple structure alignment procedure based on the protein superposition of C-alpha atoms.


