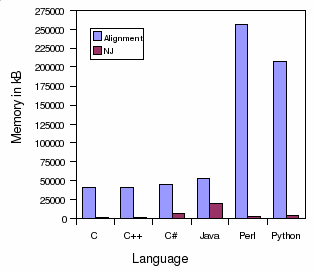Results
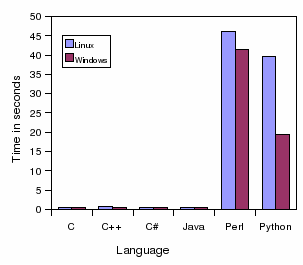
Speed Global alignment in
seconds:
This program performs a global alignment of two sequences and prints to
the standard output the aligned sequences.
|
Linux
|
Windows
|
C
|
0.38
|
0.12
|
C++
|
0.53
|
0.28
|
C#
|
0.62
|
0.37
|
Java
|
0.44
|
0.46
|
Perl
|
43.58
|
40.26
|
Python
|
23.18
|
19.21
|
|
|
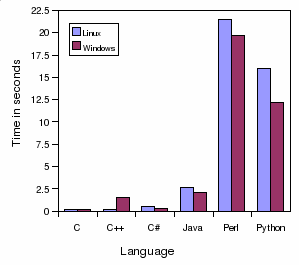
Speed Neighbor-Joining in
seconds:
This program reads DNA sequences from a FASTA file and infers an NJ
tree. The tree is printed to the standard output in the newick format.
|
Linux
|
Windows
|
|
|
Native
|
|
C
|
0.04
|
n/a
|
0.07
|
C++
|
0.12
|
n/a
|
0.15
|
C#
|
0.41
|
n/a
|
0.31
|
Java
|
2.58
|
0.71
|
2.23
|
Perl
|
11.87
|
0.29
|
11.03
|
Python
|
8.94
|
n/a
|
6.59
|
|
|
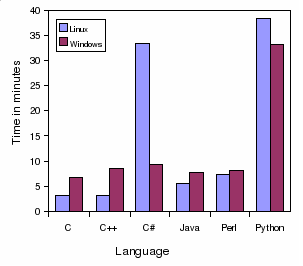
Speed BLAST parser in minutes:
This program parses a BLAST file and prints to the
standard output some information.
|
Linux
|
Windows |
|
RE
|
Tokenisation
|
RE
|
C
|
3.1
|
1.49
|
6.63
|
C++
|
3.21
|
1.45
|
8.5
|
C#
|
33.44
|
n/a |
9.21
|
Java
|
5.58
|
n/a |
7.76
|
Perl
|
7.28
|
n/a |
8.05
|
Python
|
38.42
|
n/a |
33.3
|
|
|
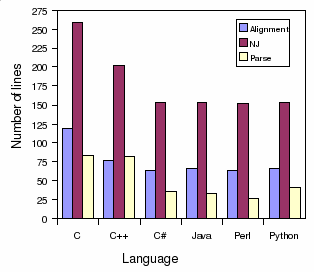
Number of line:
|
Alignment
|
NJ
|
Parser
|
C
|
119
|
240
|
82
|
C++
|
76
|
194
|
81
|
C#
|
63
|
142
|
36
|
Java
|
65
|
140
|
33
|
Perl
|
63
|
142
|
26
|
Python
|
66
|
143
|
41
|
|
|
Memory usage in kB:
|
Alignment
|
NJ
|
C
|
40892
|
1148
|
C++
|
41376
|
1680
|
C#
|
45296
|
7196
|
Java
|
52948
|
20036
|
Perl
|
256396
|
2764
|
Python
|
207728
|
3984
|
|
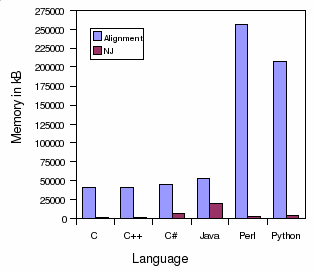
|
|