|
Unique Capabilities: FirstGlance in Jmol offers the following powerful/convenient features absent from the viewers offered by the Protein Data Bank (2025: Mol*, basic Jmol, Molmil2) as well as others such as PyMOL, ChimeraX, iCn3D:
|

|
| Initial view of Hin recombinase protein bound to DNA double helix (1ijw). |
|
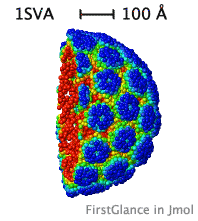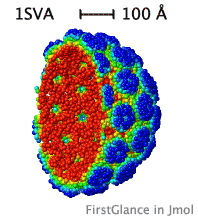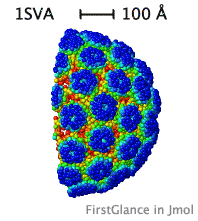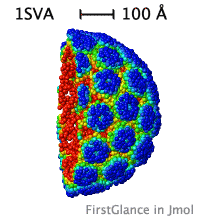
| Slabbed, simplified capsid of SV40 virus. Turn on Slab (Views tab), then, in the lower left panel, check Don't hide the back and Rotate slab. |
While the initial view is biological assembly 1, you have the option of viewing the asymmetric unit (or biological assemblies 2-N when specified). Very large assemblies (such as virus capsids) are automatically simplified: See examples of huge assemblies.

|
| Potassium channel (1R3J) showing membrane surface planes. Obtained (from OPM) via Resources tab in FirstGlance. |
-
Views Tab:
Major structural features of the
molecule are revealed. Explanatory help is shown automatically for
each view.
One-click options display secondary
structure, amino and carboxy (or 3' and 5') termini, composition
(protein, DNA, RNA, ligands, and solvent), the distributions of
hydrophobic, polar, charged amino acids, and local uncertainty
(B factor/temperature).
Non-standard residues, missing residues, and incomplete sidechains
are clearly indicated.
Centering, slab and zoom buttons make an uncluttered closeup examination easy
for any moiety of interest.
Slabbing the Hydrophobic/Polar view makes it easy to see hydrophobic cores of domains.
Save Images and Animations: Any molecular view in FirstGlance can be saved with a few clicks. It can be saved as a static image at any size desired (up to 5,000 pixels across). Or it can be saved as a rocking or spinning animation. Animations can be dropped into a slide (in Powerpoint, Google Slides, Libre Office, etc.), or displayed in web pages such as the example at right. Here are more examples of animations from FirstGlance:
-
The
Focus Box (in the Molecule Information, Views, and Tools tabs)
makes it easy to find, hide, isolate, or center items of interest.
It also has checkboxes to label every residue with its 3-letter name (or 1-letter name)
and sequence number.
- The
Button Box (in the Views and Tools tabs)
has buttons to toggle spinning, background black or white,
slabbing, showing/hiding ligands or water, zooming.
-
Tools Tab:
- A Find.. tool makes it easy to locate short sequences, and amino acids (or nucleotides) by name, sequence number, or a range of sequence numbers.
- A Hide.. tool hides any chain(s), residue(s), or atom(s) that you click. They remain hidden in other views until explicitly re-displayed.
-
An Isolate.. tool, in one click, hides everthing except one chain or ligand molecule
or range of residues, or isolates an arbitrary group of found atoms.
Isolation persists in all views until isolation is undone.
-
 The Ends.. tool lists the ends of chains, telling whether they are charged, missing,
or blocked. Clicking on one end zooms in and shows all its contacts and non-covalent interactions.
The Ends.. tool lists the ends of chains, telling whether they are charged, missing,
or blocked. Clicking on one end zooms in and shows all its contacts and non-covalent interactions.

Contacts & Non-Covalent Interactions:
Anti-Alzheimer's drug analog (*) hydrogen bonds to tyrosines in acetylcholinesterase (1gpk).
-
The Contacts.. dialog shows the non-covalent interactions to any target
moiety you select (by clicking on it, or, using Find.., by name, sequence numbers, etc.).
The non-covalent interactions
are automatically divided into seven categories:
hydrogen-bonded water, water bridges, hydrogen-bonded non-water, hydrophobic
interactions, salt bridges, cation-pi interations, and metal and miscellaneous interactions.
Checkboxes hide categories, simplifying the display.
-
Disulfide bonds are shown in the initial view, but the Tools tab offers a disulfides/S/Se
tool. It highlights and counts disulfide bonds, sulfur, and selenium atoms
(and counts cysteine, methionine, selenocysteine, selenomethionine).
- A protein salt bridge and cation-pi interaction tool highlights these, and lists them in spreadsheet-ready format. Distances and angles are easily measured with mouse clicks.
-
Resources Tab:
Here you will find plans for teaching, learning, assessment;
Alphafold for reliable prediction of structures;
Explanations with illustrated step-by-step guides provide visualization of
evolutionary conservation, atomic clashes, lipid
bilayer boundaries for integral membrane proteins, as well as
help predicting intrinsically disordered segments of a protein chain,
and sharing customized molecular views in the Proteopedia wiki.
-
Preferences Tab: You can specify
whether the molecule should spin at the beginning of a session, etc.
- Use the Where? How? Index of FirstGlance to find out whether it does something you want to do, and if so, where to find it.
In order for all information below to be evident, be sure to click Show more details in the Molecule Information Tab (and "Show More Views", "Show more tools" in those tabs).
- Interpret Rfree objectively based upon statistics in the Protein Data Bank: Better (or much better) than average, average, worse than average, or unreliable. Try 6xml.
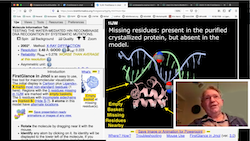
- Warn you about missing residues with "empty baskets", and mark residues with incomplete sidechains with S-, in the initial view. Tell you how many charges are missing due to missing residues, indicating the locations of missing residues with empty baskets. Tabulate the sequence numbers of missing residues for each chain. Try 1elv.
- Shows you Biological Assembly 1 initially, with options to view other Biological Assemblies (when specified) or the Asymmetric Unit.
- Automatically simplifies very large structures, such as virus capsids, making them manageable, with informative color schemes.
- Automatically colors AlphaFold and RoseTTAFold predicted structures by reliability. AlphaFold Example.
- Makes spreadsheet-ready lists of several kinds.
- Color by temperature (B factor) = "local uncertainty": relative or absolute color scheme. Identifies the chain with the lowest average temperature factor (the most reliable chain). Try 7eha: Views tab, Show more views, Local uncertainty, Show temperature of each chain.
- The Ends tool lists chain termini, telling whether they are charged, missing, or blocked. It automatically zooms in to a close-up view of a selected end, enabling you to see what it contacts, and offering an electron density map. Try 5fpk: Chain: Ends in the Molecule Information Tab, or Ends in the Tools tab.
- Indicate the presence of unusual chemical elements (those other than C, H, O, N),
and enable you to find them in the model with one click.
Try
1ihu: Click elements in the
Molecule Information Tab.
- Tell you whether all hydrogens, a subset of hydrogens, or no hydrogens are present in the model. Try 1lfa: Click 17.9% of protein atoms are hydrogen in the Molecule Information Tab.
- Display the complete molecular weight (mass) vs. the weight of the atoms present in the model. Tabulate the total weights of protein, DNA, RNA, ligands, and water in the model.
-
 Detection of protein salt bridges includes bridges involving charged protein chain termini,
and distinquishes inter-chain from intra-chain bridges. Ditto for cation-pi interactions.
Lists salt bridged atoms in a spreadsheet-ready table.
Try
9ins: With "more details" shown
(Molecule Information Tab),
click Salt Bridges in the Tools tab. Be sure to look at the Examples described
in bottom of the lower left panel.
Detection of protein salt bridges includes bridges involving charged protein chain termini,
and distinquishes inter-chain from intra-chain bridges. Ditto for cation-pi interactions.
Lists salt bridged atoms in a spreadsheet-ready table.
Try
9ins: With "more details" shown
(Molecule Information Tab),
click Salt Bridges in the Tools tab. Be sure to look at the Examples described
in bottom of the lower left panel.
- Analyze atoms with alternate locations and tabulate how many locations each such atom was assigned. Find alternate locations easily. Analyzes occupancies and summarizes them in spreadsheet-ready form. Animates alternate locations. Try 9ins: In the Molecule Information Tab, with "more details" shown, click alternate locations.
- Lists related entries in the PDB. Each can be clicked for quick examination.
This list is just above the
Focus Box in the
Molecule Information Tab. Example:
1ijw.
- Evolutionary Conservation: FirstGlance has extensive capabilities for displaying and analyzing protein PDB files pre-processed by the ConSurf Server. See Visualizing Conservation. For example, the conservation or lack of conservation of residues contacting a ligand can be visualized with the Contacts & Non-Covalent Interactions Tool of FirstGlance: see this illustration. Contacts, all salt bridges, or anything you search for with Find.. in FirstGlance can be listed spreadsheet-ready, including conservation levels. Interpreting ConSurf Results discusses additional advantages of using FirstGlance. Try 7cap_consurf1639680092_pipe.pdb. In the Tools tab, try Contacts and Protein Crosslinks. Display one isopeptide crosslink, check "Show 4.0 Å neighbors", then check "Use ConSurf Colors".
- Isopeptide bonds, thioesters, thioethers, esters, histidine-tyrosine bonds, and lysine-cysteine NOS bonds are unusual covalent crosslinks between chains. You will be alerted when they are likely present. Try 2xi9. Each putative crosslink can be visualized in one click, zooming in to see details. Examination of the electron density map is one additional click that helps to see if the putative crosslink is bona fide. See Protein Crosslinks in the Tools tab. Example cases. See also Crosslink Detection Methods and this illustrated Guide to Validation of Protein Crosslinks.
- FirstGlance has built-in Electron Density Map (or EM Density) display capability. Maps can be displayed for any small group of atoms that you identify with the Find.. tool, for ends of chains, and for protein crosslinks. For snapshots, see Electron Density Maps. For animations, see slides 11 and 12 at tinyurl.com/movingmolecules.
- You will be alerted to the presence of any of more than 30 unusual components or features. A preference setting controls whether alerts occur for most of these. These include single amino acid, dipeptide, nucleotide/nucleoside or cyclic nucleotide ligands, D amino acids, deuterium, unusual nucleotides, unusual protein crosslinks, zero or negative sequence numbers, or insertion codes. FirstGlance makes it easy to locate these in the model. Complete list of unusual components or features alerted by FirstGlance, with examples of each.
-
Crystal contacts and the unit cell (Tools tab, Show More Tools)
can each be displayed with a single click.
Use the Where? How? Index of FirstGlance to find out whether it does something you want to do, and if so, where to find it.
Customized Views. FirstGlance is designed to show you important structural features as easily as possible. Views (renderings and color schemes) are "canned" for ease of use. FirstGlance does not let you customize a view beyond its "canned" options, except for hiding or isolating portions of the structure. If you wish to customize the rendering and colors, we recommend Proteopedia.Org, which has Molecular Scene Authoring Tools. Proteopedia is the easiest place to customize a molecular scene. Scenes that you create will be immediately online for sharing with anyone. Any scene in Proteopedia can be saved as a presentation-ready animation by FirstGlance (see instructions).
FirstGlance has not yet been adapted to display mmCIF format atomic coordinate files. (Jmol has already been adapted for mmCIF files.) This means that FirstGlance cannot display some extremely large models, since the older PDB format is limited to 99,999 atoms per asymmetric unit, or a maximum molecular mass of about 1,400,000 Daltons. In July, 2025, 4.1% of the entries in the Protein Data Bank cannot be accommodated in a single PDB-format file. These are available in mmCIF format, or as multiple downloadable PDB files.
There are numerous molecular visualization software packages available. Sometimes a particular package is best suited to the job at hand. When comparing FirstGlance with another package, ask yourself whether the other package:
- Shows an equally informative initial view
(cartoon ribbons, each chain a different color,
ligands obvious, missing residues obvious).
- Displays an explanation and color keys for every view or operation that you perform,
usually providing examples for illustration and comparison with your molecule.
Has tool tips on every link and button.
Uses less technical language,
providing background information about terms
and concepts through links to external articles.
(FirstGlance help links often go to articles in
Proteopedia.Org.)
- Automatically shows
Biological Assembly 1 (functional quaternary assembly).
Many users of some other software don't realize
that they are not looking at the functional biological form of the molecule, but
rather the crystallographic
asymmetric unit (which can also be requested in FirstGlance).
Automatically
simplifies
enormous protein assemblies to create more manageable and informative views.
- Makes missing residues obvious in the initial view, tabulating them on request.
About 80% of X-ray entries in the
PDB have missing residues
(15% of NMR entries; percentages determined May, 2016). Labels residues with
incomplete sidechains.
38% of X-ray entries have them.
Most of the time users
don't realize that anything is missing.
- Gives the full name of each ligand or non-standard residue. Clicking locates each
in the model.
- Has one-click renderings and color schemes to show you
secondary structure (with percentages),
spacefilling solid (van der Waals) renderings,
the amino and carboxy termini (or 5' and 3' for nucleic acids), distribution
of hydrophobic/polar residues on the surface and in the core of the protein,
and distribution
of charges with counts and resources for
calculating pI and net charge at various pH's.
(Views tab.)
- Enables you to find anything of interest in the 3D model: sequence motifs,
sequence numbers or ranges, particiular amino acids or nucleotides, etc. etc.
- Lets you isolate with one click
any single chain, residue, ligand, or arbitrary subset of atoms
that you have tagged with Find, for closer
examination. Also you can hide chains, residues, ligands easily by clicking on them.
- Displays key information from the header of the
PDB file in readable form, with extensive explanations (the Molecule
Information Tab in FirstGlance).
- Informs you about the quality of the model by objectively grading free R
as better than average, average, worse than average, or unreliable, and explaining
why free R is crucial and how it is graded.
- Tells you the sequences of the crystallized fragment and of the full-length protein,
and which chains in the model are identical in sequence.
And you can label each residue by sequence number and/or residue name, or click on it
to report the number and name. Offers
instructions for making a sequence alignment
between the experimental (typically crystallized) sequence and the full-length genomic sequence,
and provides an easy way to view sequence alignments
(MSAReveal.Org).
- Displays disulfide bonds, sulfur-containing residues,
selenium-containing residues and statistics.
Salt bridges and cation-pi interactions.
(Tools tab.)
- Offers a tool to see all atoms/residues contacting any moiety you choose -- including all
non-covalent interactions.
(Tools tab.)
- Provides a link to view the abstract of the publication at PubMed.
- Tabulates an analysis of alternate locations and occupancies
with links to view subsets of interest.
(Molecule Information Tab)
- Shows you Crystal contacts with examples of how they might affect the conformation
of the protein in the crystal. Also the unit cell.
(Tools tab.)

Enolase (4enl; a glycolytic enzyme) evolutionary conservation from ConSurf. Catalytic pocket is highly conserved. More.. 
- Provides an
Introduction to Evolutionary Conservation as well as guidance for those
with experience. FirstGlance can directly display proteins colored by evolutionary
conservation (in collaboration with the
ConSurf Server). This enables you to see the conservation of residues
contacting ligand, involved in protein-protein interactions, as well as identifying
functional regions based on their high level of conservation.
- Offers assistance for using external resources to see atomic clashes (MolProbity),
predict intrinsically disordered protein, see lipid bilayer boundaries on
integral membrane proteins (Orientations of Proteins in Membranes).
(Resources tab.)
- Indicates unusual chemical elements in the model, and locates them.
- Provides a
master index
where you can find how to access a particular tool or
capability.
- Defines and explains the technical terms it uses (such as "metal", "carbohydrate", "non-standard residue", "biological unit", "asymmetric unit", "free R").
Jmol is developed separately by its own open-source volunteer team, primarily by Robert M. Hanson. FirstGlance in Jmol has been developed largely by Eric Martz, with much-appreciated help from many others. Others are free to get involved since it is an open-source project. FirstGlance in Jmol uses the Creative Commons Attribution-Noncommercial-Share Alike 3.0 License, while Jmol itself uses the GNU Lesser General Public License.


