Format Sequence data: If your data is in the form of DNA sequence you can use this utility tool to format your data. Your sequence should be as Fasta format.
You need to have two files one with negative examples and the others with positive examples.
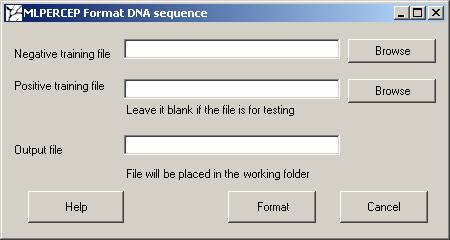
Click on Format Sequence Data and you will get the below shown window.
eg:
>Seq1 Hypothetical gene
ATGCGTA
>Seq2 Kinase
TTCTAAC
And so on. Here the first word will be taken as the sequence name. For the above example it will be Seq1 and Seq2
Negative training file: Upload the file with sequences which belong to one group. Here it will be assigned 0.
Positive training file: Upload the file with sequences which belong to the other group. Here it will be assigned 1.
The sequence are numerically transferred as
Ač0 0
Tč0 1
Gč1 0
Cč1 1
Unidentified bases will be assigned 0 0
Its very important that the length of sequences are equal.
Output file: Specify the output file name.
Click on format. You will see a dos window which will tell you once your data is formatted. For large datasets it might take some time. Upload this file in the training program.
For testing purpose you need only one file. Leave the field Positive training file as empty. The system will consider that the formatting is done for the testing purpose. It wont put any expected output and in the testing program you need to uncheck “Second row contain target”.