Bldb concept
From Bioinformatics.Org Wiki
![]() BioLegato#bldb return to bioLegato
BioLegato#bldb return to bioLegato
Contents |
Rationale
Bldb is intended as a database client for querying and for editing database content. Ideally, the database should be any SQL database as a back end.
Why do we need another database client?
There are a number of GUI database clients
All of the DB clients I looked at are oriented toward database managers. The end user has to know SQL and conceptualize data in the context of SQL.
What we want is a GUI that is model oriented - that presents the data in a form familiar to the end user.
While we don't want to inflict these GUIs on the end user, any of these engines might be suitable as a backend. In particular, hsqldb can be accessed through a Java API, which might be useful.
ACeDB is a good model for client behavior. Its strengths include an abilty to do either simple queries or complex queries without having to know SQL. As well, ACeDB is able to display individual records (objects) as popup windows in which individual data fields can be clicked on, resulting in the records they point to popping up. Complex queries are done using Query by Example. There are two main reasons for replacing ACeDB. First, the code is old C code and is no longer being developed or supported. The second is that ACeDB includes the client and database engine in a single program, and the ACeDB database engine doesn't scale well to large databases.
ACeDB has some unique features:
- Can display or edit an individual record in a separate window
- Can execute external programs eg. launch an image viewer or open a web page
- Query builder
- Query by exampe
- Table builder
bltable vs. bldb - Superficially, there seem to be many advantages to implementing bldb by extending bltable. That would require re-casting the existing String type, used for the contents of each cell, into Object. This my be a very good investment, in the long run, because improvements in bltable would automatically be inherited by bldb..
Implementation Strategy
The main idea is to evolve birchdb in such a way that components are created in such an order that we never break the important functionalities.
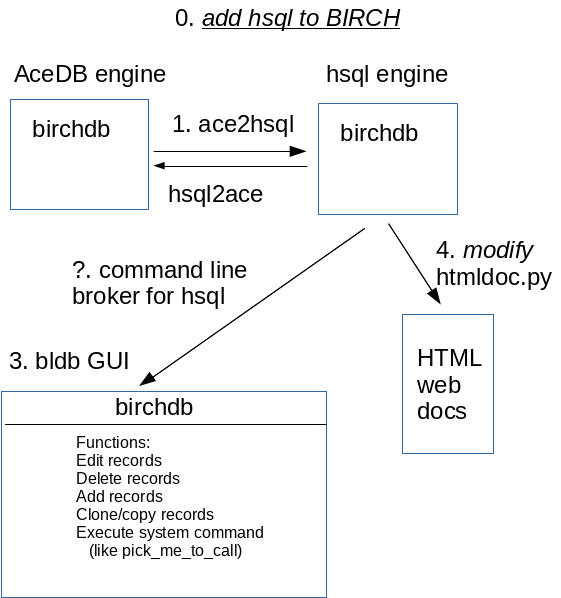
0. add hsql to BIRCH
We do this step first because it doesn't break anything, and it is critical for all later steps.
1. ace2hsql
ace2hsql is a script or program that dumps the contents of the birchdb database (both core and local) to an ACeDB flatfile (.ace), and updates the existing birchdb copy in the hsql databse. At this point we are still doing all actual work on the birchdb database using ACeDB, but the contents of birchdb get mirrored to the hsql database.
This requires that we write an SQL schema that maps to the schema for birchdb. Then the problem boils down to writing a script that can read a flat file dump of birchdb, and populate the SQL database with the corresponding data objects.
2. modify htmldoc.py to extract records from birchdb
This step assumes an up to date mirror of birchdb is found in the hsql database. The existing htmldoc.py script knows how to read the .acefile from the ACeDB version of birchdb. We need to teach htmldoc.py how to also do its input from the hsql birchdb.
At this point, we are still doing all changes to the ACeDB copy of the database, but mirroring changes to hsql.
3. commandline broker for hsql
The commandline broker for hsql knows how to do transactions with the hsql database at the command line. The idea is that its input is simple. You shouldn't need to know how to write SQL to run this.
Commands include:
- Edit records
- Delete records
- Add records
- Clone/copy records
4. bldb front end for database
When this step is complete, we will have a fully-functional birchdb, using the hsql database engine and BioLegato client as a GUI.
Functions:
- Query by example
- Edit records
- Delete records
- Add records
- Clone/copy records
Individual records can be viewed, as in ACeDB (or the old HyperCard).