Main.Tutorial HistoryHide minor edits - Show changes to markup September 15, 2010, at 03:39 PM
by -
Changed lines 49-50 from:
Comgratulations!!ChemChains is now installed and ready for your first simulation! to:
Congratulations!! ChemChains is now installed and ready for your first simulation! December 05, 2009, at 04:26 PM
by -
Changed lines 294-295 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt
-ispecs inFiles/exts/CChain/specs/sampleSpecs.txt to:
[@./ChemChains -ilogic inFiles/CChain/logic/sampleNet.txt
-ispecs inFiles/CChain/specs/sampleSpecs.txt Changed lines 355-357 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt
-ispecs inFiles/exts/CChain/specs/sampleSpecs.txt -patterns -isettings inFiles/exts/exts/Patterns/settings.txt to:
[@./ChemChains -ilogic inFiles/CChain/logic/sampleNet.txt
-ispecs inFiles/CChain/specs/sampleSpecs.txt -patterns -isettings inFiles/exts/Patterns/settings.txt December 05, 2009, at 04:23 PM
by -
Changed lines 269-271 from:
-itables inFiles/exts/FileConverter/sampleNet@ -o outputFileName.txt] to:
-itables inFiles/exts/FileConverter/sampleNet -o outputFileName.txt@] December 05, 2009, at 04:22 PM
by -
Changed lines 268-270 from:
-iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet@] to:
-inodelist inFiles/exts/FileConverter/sampleNodeList.txt -itables inFiles/exts/FileConverter/sampleNet@ -o outputFileName.txt] April 20, 2009, at 02:03 PM
by -
Changed line 362 from:
ChemChains can be run with randomly generated conditions for every simulation. To run ChemChains with random initial conditions enabled, use the '-rand_init' flag. to:
ChemChains can be run with randomly generated conditions for every simulation. To run ChemChains with random initial conditions enabled, use the '-randInit' flag. February 28, 2009, at 11:11 AM
by -
Changed line 144 from:
Bool:B:True:A,C:FF,FT,TT
to:
Bool:B:True:C,A:FF,FT,TT
February 28, 2009, at 12:29 AM
by -
Changed line 355 from:
-ispecs inFiles/exts/CChain/logic/sampleNet.txt to:
-ispecs inFiles/exts/CChain/specs/sampleSpecs.txt February 28, 2009, at 12:28 AM
by -
Changed line 294 from:
-ispecs inFiles/exts/CChain/logic/sampleNet.txt to:
-ispecs inFiles/exts/CChain/specs/sampleSpecs.txt February 28, 2009, at 12:26 AM
by -
Changed line 275 from:
-ispecs inFiles/exts/CChain/logic/sampleNet.txt to:
-ispecs inFiles/exts/CChain/specs/sampleSpecs.txt November 17, 2008, at 11:36 AM
by -
Changed lines 44-46 from:
When Cygwin is launched you will begin in the default directory, C:\cygwin\~. to:
November 17, 2008, at 11:33 AM
by -
Changed line 34 from:
to:
Changed line 37 from:
to:
Changed line 44 from:
to:
Changed lines 47-51 from:
to:
Comgratulations!!ChemChains is now installed and ready for your first simulation! November 17, 2008, at 11:16 AM
by -
Changed line 34 from:
to:
November 17, 2008, at 11:13 AM
by -
Changed line 34 from:
to:
Changed lines 40-41 from:
to:
Added lines 44-45:
November 17, 2008, at 10:54 AM
by -
Changed lines 39-40 from:
to:
November 17, 2008, at 10:45 AM
by -
Changed line 38 from:
to:
November 17, 2008, at 10:43 AM
by -
Changed line 38 from:
to:
November 17, 2008, at 10:40 AM
by -
Changed lines 36-37 from:
to:
November 17, 2008, at 10:38 AM
by -
Changed line 35 from:
to:
November 17, 2008, at 10:37 AM
by -
Changed line 35 from:
to:
November 14, 2008, at 11:16 AM
by -
Added line 32:
November 14, 2008, at 11:14 AM
by -
Changed line 31 from:
to:
November 14, 2008, at 11:13 AM
by -
Changed lines 40-41 from:
Running Cygwin
to:
Running Cygwin & Setting up ChemChains
November 14, 2008, at 11:11 AM
by -
Changed line 21 from:
to:
November 14, 2008, at 11:10 AM
by -
Changed line 21 from:
to:
November 14, 2008, at 11:08 AM
by -
Changed lines 15-16 from:
For example, you can download and install the freely available cygwin from here. to:
You can use a freely available Cygwin (see below). October 31, 2008, at 12:01 PM
by -
Changed lines 44-45 from:
to:
October 21, 2008, at 05:27 PM
by -
Changed line 229 from:
Flag to Specify the Network Descriptor File
to:
Flag to Specify the Simulation Specification File
October 21, 2008, at 05:26 PM
by -
Changed lines 230-231 from:
./ChemChains -ilogic [network descriptor filename]
to:
./ChemChains -ispecs [simulation specification filename]
October 21, 2008, at 05:26 PM
by -
Added lines 229-231:
Flag to Specify the Network Descriptor File
./ChemChains -ilogic [network descriptor filename]
October 21, 2008, at 05:23 PM
by -
Changed lines 144-145 from:
Once finished, save the file as to:
Once finished, save the file as Added line 148:
\\ October 21, 2008, at 05:22 PM
by -
Added line 111:
Added lines 146-147:
Flag to Specify the Network Descriptor File
./ChemChains -ilogic [network descriptor filename]
October 21, 2008, at 05:19 PM
by -
Changed line 68 from:
to:
Changed line 70 from:
to:
October 21, 2008, at 05:17 PM
by -
Changed lines 127-128 from:
to:
October 21, 2008, at 05:16 PM
by -
Changed lines 127-128 from:
to:
October 21, 2008, at 05:15 PM
by -
Added line 113:
\\ Added lines 125-126:
These activation mechanisms can be expressed as individual truth tables (Figure 2). Deleted lines 128-130:
These activation mechanisms can be expressed as individual truth tables (Figure 2). October 21, 2008, at 05:15 PM
by -
Deleted line 112:
\\ October 21, 2008, at 05:14 PM
by -
Added line 127:
Changed lines 151-152 from:
\\(The following example of a simple simulation specification file is included with the current ChemChains release and can be found in "inFiles/CChain/specs/sampleSpecs.txt" file in the ChemChains directory) to:
(The following example of a simple simulation specification file is included with the current ChemChains release and can be found in "inFiles/CChain/specs/sampleSpecs.txt" file in the ChemChains directory) October 21, 2008, at 05:13 PM
by -
Added lines 150-151:
\\(The following example of a simple simulation specification file is included with the current ChemChains release and can be found in "inFiles/CChain/specs/sampleSpecs.txt" file in the ChemChains directory) October 21, 2008, at 05:12 PM
by -
Changed lines 113-114 from:
(The following example of a simple toy network is available with the current ChemChains release and can be found in "inFiles/CChain/logic/sampleNet.txt" file in the ChemChains directory) to:
October 21, 2008, at 05:11 PM
by -
Deleted lines 52-54:
Changed lines 112-113 from:
(The following example of a simple toy network is available with the current ChemChains release and can be found in "inFiles/CChain/logic/sampleNet.txt" file in the ChemChains directory) to:
October 21, 2008, at 05:10 PM
by -
Changed lines 115-116 from:
to:
(The following example of a simple toy network is available with the current ChemChains release and can be found in "inFiles/CChain/logic/sampleNet.txt" file in the ChemChains directory) October 21, 2008, at 04:58 PM
by -
Changed lines 27-28 from:
to:
October 21, 2008, at 04:57 PM
by -
Changed lines 26-27 from:
to:
October 21, 2008, at 04:55 PM
by -
Changed line 21 from:
to:
October 21, 2008, at 04:54 PM
by -
Added lines 19-23:
Download
October 20, 2008, at 02:29 PM
by -
Changed line 36 from:
When Cygwin is started the default directory is C:\cygwin\~. to:
When Cygwin is launched you will begin in the default directory, C:\cygwin\~. October 20, 2008, at 02:28 PM
by -
Changed lines 31-32 from:
run ChemChains the following packages are necessary from the Developer packages IN ADDITION TO the ones preselected by Cygwin: binutils, gcc-c++, and make packages to:
Changed lines 35-42 from:
There are a couple useful commands for working in a Unix environment, to:
When Cygwin is started the default directory is C:\cygwin\~.
Some basic Unix commands Changed lines 47-54 from:
When you start Cygwin it will start you into C:\cygwin\~, you need to cd .. to get to C:\cygwin, when you get to the directory that you put the ChemChains binary into you can use the command make to compile it. When it finishes it creates the executable ChemChains, to run it it use the command ./ChemChains along with the flags you want to use. You can use ./ChemChains -h to view flag usage documentation. to:
October 20, 2008, at 02:18 PM
by -
Changed lines 25-43 from:
First go here and download the file Install.exe, when you run it it will open the Cygwin installation wizard. If you don't have any cygwin packages downloaded allready select Install From Internet and hit next. It will prompt you for a root directory with the default C:\cygwin. You can use another directory if you want but I would advise against using C:\. It will then prompt you for a package directory, this is where it will keep the installation files for the packages you install, I usually use the same directory as I selected for root. Click next and it will prompt you for your connection type, select it and hit next, then select a mirror to download packages from. Then it will bring up a package list interface. To compile and run ChemChains you will need the binutils, gcc-c++, and make packages from the Developer packages in addition to all of the packages that Cygwin will automatically select. After you select the packages that you want hit next and the installer will download and install the packages you've selected. If anytime in the future you find that you need a new package installed you can re-run the Cygwin installer and select the package(s) you need and it will install them for you. to:
run ChemChains the following packages are necessary from the Developer packages IN ADDITION TO the ones preselected by Cygwin: binutils, gcc-c++, and make packages
September 30, 2008, at 03:36 PM
by -
Changed lines 48-50 from:
to:
September 30, 2008, at 03:22 PM
by -
Changed lines 24-26 from:
to:
Installing Cygwin
First go here and download the file Install.exe, when you run it it will open the Cygwin installation wizard. If you don't have any cygwin packages downloaded allready select Install From Internet and hit next. It will prompt you for a root directory with the default C:\cygwin. You can use another directory if you want but I would advise against using C:\. It will then prompt you for a package directory, this is where it will keep the installation files for the packages you install, I usually use the same directory as I selected for root. Click next and it will prompt you for your connection type, select it and hit next, then select a mirror to download packages from. Then it will bring up a package list interface. To compile and run ChemChains you will need the binutils, gcc-c++, and make packages from the Developer packages in addition to all of the packages that Cygwin will automatically select. After you select the packages that you want hit next and the installer will download and install the packages you've selected. If anytime in the future you find that you need a new package installed you can re-run the Cygwin installer and select the package(s) you need and it will install them for you. Running Cygwin
There are a couple useful commands for working in a Unix environment,
When you start Cygwin it will start you into C:\cygwin\~, you need to cd .. to get to C:\cygwin, when you get to the directory that you put the ChemChains binary into you can use the command make to compile it. When it finishes it creates the executable ChemChains, to run it it use the command ./ChemChains along with the flags you want to use. You can use ./ChemChains -h to view flag usage documentation. August 23, 2008, at 09:56 AM
by -
Changed lines 183-184 from:
To create a delay node, use the following syntax: to:
To create a sustain node, use the following syntax: July 27, 2008, at 01:41 AM
by -
Added lines 61-62:
July 27, 2008, at 01:39 AM
by -
Changed lines 183-184 from:
Sustain:name:initial value:parent node:duration (in iteration units)
to:
Sustain:name:initial value:parent node:duration
July 27, 2008, at 01:38 AM
by -
Changed lines 294-295 from:
default:lower_limit1 higher_limit1 code1,lower_limit2 higher_limit2 code2
to:
default:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2 Changed lines 302-303 from:
NodeName:lower_limit1 higher_limit1 code1,lower_limit2 higher_limit2 code2
to:
NodeName:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2 July 26, 2008, at 11:10 PM
by -
Changed line 37 from:
Table 1: List of flags available in the calculation mode. to:
Table 1: List of currently available flags. Changed line 52 from:
to:
Changed lines 257-267 from:
to:
July 26, 2008, at 11:02 PM
by -
Added lines 58-60:
Changed lines 70-73 from:
to:
July 26, 2008, at 11:00 PM
by -
Changed lines 31-32 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. All flags available in the most recent version of ChemChains are described in Table 1. Deleted line 34:
All flags available in the most recent version of ChemChains are described in Table 1. July 26, 2008, at 11:00 PM
by -
Added lines 35-79:
All flags available in the most recent version of ChemChains are described in Table 1.
Changed lines 246-247 from:
To run ChemChains in the calculation mode, use the '-calc' flag. Other flags available within the calculation mode are listed in Table 2. to:
To run ChemChains in the calculation mode, use the '-calc' flag. You can run ChemChains (using our previously created network descriptor and simulation specification files) in the calculation mode in the most basic way (without the use of any extensions) as follows: ./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -n 5 -calc myFirstCalcSimulation This command will simulate our toy network five times and will save all output in CCOutput/stats/myFirstCalcSimulation folder. The following table describes the structure and contents of the output folder. Changed line 260 from:
Table 2: List of flags available in the calculation mode. to:
Table 3: Contents of output generated during calculation mode. Deleted lines 270-292:
You can run ChemChains (using our previously created network descriptor and simulation specification files) in the calculation mode in the most basic way (without the use of any extensions) as follows: ./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -n 5 -calc myFirstCalcSimulation This command will simulate our toy network five times and will save all output in CCOutput/stats/myFirstCalcSimulation folder. The following table describes the structure and contents of the output folder.
July 26, 2008, at 10:19 PM
by -
Changed lines 175-176 from:
Save the file as to:
Save the file as Changed lines 179-181 from:
./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet to:
./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet Changed lines 249-250 from:
default:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2
to:
default:lower_limit1 higher_limit1 code1,lower_limit2 higher_limit2 code2
Changed lines 256-257 from:
NodeName:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2
to:
NodeName:lower_limit1 higher_limit1 code1,lower_limit2 higher_limit2 code2
July 26, 2008, at 10:17 PM
by -
Changed lines 94-95 from:
to:
Note: All parameters are case sensitive. July 26, 2008, at 10:17 PM
by -
Changed lines 94-95 from:
Note: All parameters are case sensitive. to:
July 26, 2008, at 10:17 PM
by -
Changed lines 89-92 from:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below) to:
Input:name:initial/default value:random(R)/fixed(F): noise(N)/fixed/(F):time to be introduced:dose (in percentage): duration of the dosage July 26, 2008, at 10:16 PM
by -
Changed lines 56-58 from:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) to:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) July 26, 2008, at 10:15 PM
by -
Changed lines 146-147 from:
[@Output:output name:initial value:node name]
to:
Output:output name:initial value:node name
July 26, 2008, at 10:13 PM
by -
Changed line 93 from:
Note: Each input needs to be declared on a new line. to:
Note: Each input needs to be declared on a new line. Changed lines 134-135 from:
[
Delay:name:initial value:parent node:iterations to wait]
to:
Delay:name:initial value:parent node:iterations to wait
Changed lines 139-140 from:
[
Sustain:name:initial value:parent node:duration (in iteration units)]
to:
Sustain:name:initial value:parent node:duration (in iteration units)
Changed lines 146-147 from:
to:
[@Output:output name:initial value:node name]
July 26, 2008, at 10:12 PM
by -
Changed lines 41-44 from:
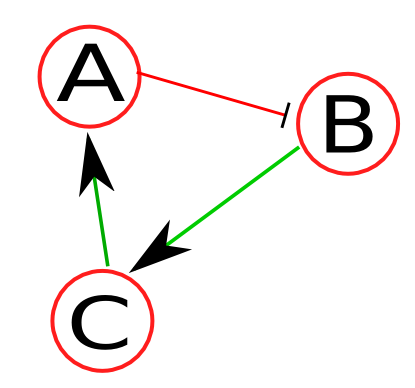
 Figure 1: Sample network. As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, C, and external input IN1, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows: to:
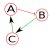
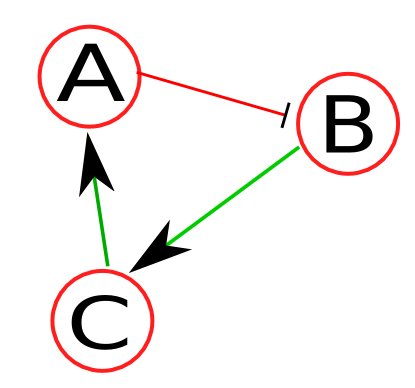
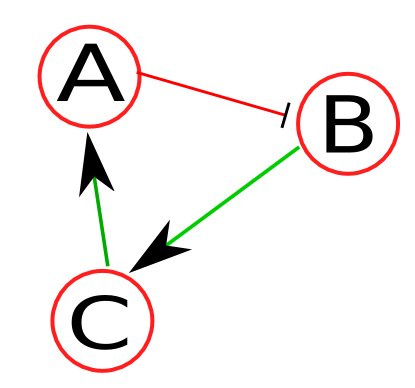
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, C, and external input IN1, where green line (with an arrow) denotes positive regulation (e.g., activation) and red line (with a stop line) denotes negative regulation (e.g., inhibition). Because we're dealing with a boolean network, each node has an activation mechanism (represented by truth tables) associated with it. As an example, the activation mechanisms for the nodes of this network are as follows: Changed lines 50-55 from:
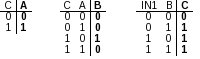 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Fig. 1 These activation mechanisms can be expressed as individual truth tables (Figure 2). Once we have the network and truth tables established, we can create our network descriptor file using these tables. To create the file, the following syntax needs to be used: to:
These activation mechanisms can be expressed as individual truth tables (Figure 2). Once the network and truth tables are established, they can be used to create the network descriptor file. Use the following syntax to create the file: Changed lines 81-84 from:
For our example, we'll set the total time to 20 iterations and transient time to 10 as follows: RunTime:10:20
to:
For our example, we'll set the total time to 100 iterations and transient time to 70 as follows: RunTime:70:100
Changed lines 93-94 from:
Note: Each input needs to be declared on a new line to:
Note: Each input needs to be declared on a new line. Note: All parameters are case sensitive. Changed line 97 from:
to:
Added lines 128-130:
Note: The name of the delay, sustain, and output nodes has to differ from the names of their inputs. Changed lines 132-135 from:
[Needs content]
to:
To create a delay node, use the following syntax: [
Delay:name:initial value:parent node:iterations to wait]
Changed lines 137-140 from:
[Needs content]
to:
To create a delay node, use the following syntax: [
Sustain:name:initial value:parent node:duration (in iteration units)]
Changed lines 142-143 from:
[Needs content]
to:
Output nodes are nodes that will be printed out via ChemChains' visual mode. To create a new output node, use the following syntax: Changed lines 149-150 from:
Depending on which mode ChemChains is run in, different output will be generated. In general, both the visual and calculation modes will create and save their output in a directory called 'CCOutput' that will be created (if it already doesn't exist) in a directory directly above the directory with ChemChains software. Refer to the Running Modes section on detailed description of output generated by ChemChains. to:
Depending on which mode ChemChains is run in, different output will be generated. In general, both the visual and calculation modes will create and save their output in a directory called 'CCOutput' that will be created (if it already doesn't exist) in a directory directly above the directory with ChemChains software. Refer to the Running Modes section on detailed description of output generated by ChemChains. Deleted line 151:
Changed lines 159-160 from:
Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. to:
Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. Changed lines 170-174 from:
A 1 0
B 2 0
C 3 0
IN1 IN101 1
to:
A 1
B 2 1
C 3 1
IN1 IN4
July 26, 2008, at 08:22 PM
by -
Changed lines 36-37 from:
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file. to:
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file (additional input files may be required for individual running modes. Changed line 134 from:
to:
Changed lines 137-138 from:
to:
July 26, 2008, at 08:20 PM
by -
Changed lines 31-32 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. Changed line 35 from:
to:
July 26, 2008, at 08:19 PM
by -
Changed lines 36-38 from:
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file. Network Descriptor
to:
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file. Network Descriptor
Changed line 72 from:
Simulation Specification file
to:
July 25, 2008, at 04:13 PM
by -
Changed line 3 from:
to:
July 25, 2008, at 04:11 PM
by -
Changed lines 3-5 from:
to:
July 25, 2008, at 03:59 PM
by -
Changed lines 3-6 from:
to:
Table of Contents
July 25, 2008, at 03:56 PM
by -
Changed lines 4-5 from:
to:
July 25, 2008, at 03:50 PM
by -
Changed lines 102-103 from:
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
to:
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
Changed lines 105-106 from:
to:
July 25, 2008, at 03:49 PM
by -
Added line 106:
July 25, 2008, at 03:49 PM
by -
Changed lines 2-3 from:
font-size=smaller bgcolor=#e0e0ff% to:
July 25, 2008, at 03:46 PM
by -
Changed lines 2-3 from:
to:
font-size=smaller bgcolor=#e0e0ff% July 25, 2008, at 03:46 PM
by -
Changed lines 102-103 from:
%div >advanced apply=div%< Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
to:
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
Changed line 105 from:
%div > apply=div%<
to:
July 25, 2008, at 03:45 PM
by -
Changed lines 2-3 from:
to:
Changed lines 102-103 from:
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following: to:
%div >advanced apply=div%< Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
Changed line 105 from:
to:
%div > apply=div%<
July 25, 2008, at 03:44 PM
by -
Changed lines 2-3 from:
% to:
July 25, 2008, at 03:43 PM
by -
Changed lines 2-3 from:
to:
% Changed lines 100-101 from:
Input:IN1:True:F:F:1:10:20
to:
Input:IN1:True:F:F:1:10:99
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following: Input:IN1:True:R:F:1:0-10:29,30:11-50:39,71:51-100:29
July 25, 2008, at 03:32 PM
by -
Changed lines 242-246 from:
-n 5 -calc myFirstCalcSimulation @] to:
-n 5 -calc myFirstCalcSimulation @] Random Initial Conditions
ChemChains can be run with randomly generated conditions for every simulation. To run ChemChains with random initial conditions enabled, use the '-rand_init' flag. July 25, 2008, at 03:28 PM
by -
Changed lines 71-72 from:
to:
Changed lines 79-80 from:
to:
Changed lines 101-103 from:
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level at time t will be 10%+/-5.
to:
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level at time t will be 10+/-5%.
Changed line 110 from:
to:
Changed line 116 from:
to:
Changed line 118 from:
to:
Changed line 120 from:
to:
July 25, 2008, at 03:25 PM
by -
Changed lines 102-103 from:
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level, at time t, will be 10%+/-5.
to:
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level at time t will be 10%+/-5.
July 25, 2008, at 03:23 PM
by -
Changed lines 30-31 from:
To simulate logical networks with ChemChains, two input files are required: the network descriptor and a simulation specification file. to:
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file. July 25, 2008, at 03:12 PM
by -
Changed lines 1-2 from:
to:
Added lines 55-56:
Note: The declaration of each node is on a new line. Changed line 61 from:
Note: that the declaration of each node is on a new line. to:
Changed lines 81-82 from:
Syntax to declare an input (note that each input needs to be declared on a new line: to:
Syntax to declare an input: Added lines 87-88:
Note: Each input needs to be declared on a new line Added lines 143-144:
Note: If the initial value is False (i.e., 0 ) we can leave the third column empty. Changed line 150 from:
Note that if the initial value is False (i.e., 0 ) we can leave the third column empty.
to:
Changed lines 165-166 from:
Note: Make sure that you have created at least one output node in the simulation specification file! to:
Note: Make sure that you have created at least one output node in the simulation specification file! Changed lines 171-172 from:
Note: When running in the visual mode, ChemChains can run only one simulation at a time. To run multiple simulations in an automated fashion, run ChemChains in calculation mode. to:
Note: When running in the visual mode, ChemChains can run only one simulation at a time. To run multiple simulations in an automated fashion, run ChemChains in calculation mode. July 25, 2008, at 03:06 PM
by -
Added lines 1-2:
Changed line 59 from:
Note that the declaration of each node is on a new line. to:
Note: that the declaration of each node is on a new line. July 25, 2008, at 03:00 PM
by -
Changed lines 1-2 from:
to:
July 25, 2008, at 02:59 PM
by -
Changed lines 1-2 from:
to:
Changed line 225 from:
List of nodes to be analyzed'''
to:
July 25, 2008, at 02:59 PM
by -
Changed line 225 from:
to:
List of nodes to be analyzed'''
July 25, 2008, at 02:58 PM
by -
Changed lines 1-2 from:
to:
July 25, 2008, at 02:57 PM
by -
Changed lines 1-2 from:
to:
July 25, 2008, at 02:53 PM
by -
Changed line 210 from:
to:
Changed line 225 from:
to:
July 25, 2008, at 02:52 PM
by -
Changed line 210 from:
Settings File
to:
Changed line 225 from:
List of nodes to be analyzed
to:
July 25, 2008, at 02:51 PM
by -
Changed line 225 from:
to:
List of nodes to be analyzed
July 25, 2008, at 02:50 PM
by -
Changed lines 1-2 from:
to:
Added line 9:
July 25, 2008, at 02:40 PM
by -
Deleted lines 0-12:
Changed line 209 from:
to:
Settings File
July 25, 2008, at 02:40 PM
by -
Deleted line 15:
Changed line 221 from:
to:
July 25, 2008, at 02:38 PM
by -
Added lines 14-15:
July 25, 2008, at 02:31 PM
by -
Added line 5:
Changed lines 220-221 from:
Settings File
to:
Changed line 236 from:
List of nodes to be analyzed
to:
July 25, 2008, at 02:25 PM
by -
Changed lines 20-21 from:
to:
Changed line 25 from:
Windows
to:
Windows
Changed lines 29-30 from:
to:
Changed line 37 from:
to:
Changed line 40 from:
Network Descriptor
to:
Network Descriptor
Changed line 72 from:
Simulation Specification file
to:
Simulation Specification file
Changed line 127 from:
to:
Changed lines 130-131 from:
to:
Changed line 134 from:
File Conversion Mode
to:
File Conversion Mode
Changed line 160 from:
Visual Mode
to:
Visual Mode
Changed line 175 from:
Calculation Mode
to:
Calculation Mode
Changed line 216 from:
Patterns Analysis
to:
Patterns Analysis
Changed line 219 from:
Settings File
to:
Settings File
Changed line 234 from:
List of nodes to be analyzed
to:
List of nodes to be analyzed
July 25, 2008, at 02:24 PM
by -
Changed line 13 from:
(:num;) to:
July 24, 2008, at 12:25 PM
by -
Changed line 5 from:
to:
Changed lines 7-11 from:
to:
July 24, 2008, at 12:15 PM
by -
Changed lines 5-11 from:
to:
July 24, 2008, at 10:44 AM
by -
Changed lines 43-44 from:
 Figure 1: Sample network. to:
 Figure 1: Sample network. Changed lines 52-53 from:
 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Fig. 1 to:
 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Fig. 1 July 24, 2008, at 10:41 AM
by -
Changed lines 45-47 from:
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows:
to:
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, C, and external input IN1, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows:
Changed lines 49-50 from:
to:
July 24, 2008, at 10:36 AM
by -
Changed lines 67-68 from:
Once finished, save the file as to:
Once finished, save the file as The next section describes how to create the simulation specification file. July 24, 2008, at 10:35 AM
by -
Changed lines 67-68 from:
Once finished we can save the file as to:
Once finished, save the file as July 24, 2008, at 10:33 AM
by -
Changed lines 63-65 from:
Bool:B:False:A,C:FF,FT,TT
Bool:C:False:B:T
to:
Bool:B:True:A,C:FF,FT,TT
Bool:C:True:IN1,B:FF
July 24, 2008, at 10:30 AM
by -
Changed lines 51-52 from:
 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Figure 1 to:
 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Fig. 1 July 24, 2008, at 10:29 AM
by -
Changed lines 51-52 from:
 Figure 2: Truth tables for nodes A, B, and C. to:
 Figure 2: Truth tables for nodes A, B, and C, representing the sample network in Figure 1 July 23, 2008, at 05:22 PM
by -
Added line 79:
Added line 108:
Added line 110:
Added line 115:
Changed line 160 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt
Changed line 192 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt
Changed line 236 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt
July 23, 2008, at 05:17 PM
by -
Changed lines 85-87 from:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage(explanation of each option below) to:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below) July 23, 2008, at 05:16 PM
by -
Changed lines 57-59 from:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) to:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) Changed lines 155-156 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt Changed lines 187-188 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt July 23, 2008, at 05:14 PM
by -
Changed lines 229-230 from:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -patterns -isettings inFiles/exts/exts/Patterns/settings.txt -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt to:
[@./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -patterns -isettings inFiles/exts/exts/Patterns/settings.txt -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt July 23, 2008, at 05:13 PM
by -
Changed lines 149-150 from:
./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet
to:
./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet July 23, 2008, at 05:13 PM
by -
Changed lines 57-58 from:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma)
to:
Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) July 23, 2008, at 05:11 PM
by -
Changed lines 153-154 from:
to:
./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -vis myFirstSimulation.txt July 23, 2008, at 05:10 PM
by -
Changed lines 183-184 from:
to:
./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -n 5 -calc myFirstCalcSimulation July 23, 2008, at 05:10 PM
by -
Changed lines 84-85 from:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below)
to:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage(explanation of each option below) July 23, 2008, at 05:07 PM
by -
Changed lines 224-225 from:
./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt [[<<]] -patterns -isettings inFiles/exts/exts/Patterns/settings.txt -n 5 -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt -calc myFirstCalcSimulation to:
./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt -patterns -isettings inFiles/exts/exts/Patterns/settings.txt -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt -n 5 -calc myFirstCalcSimulation July 23, 2008, at 05:07 PM
by -
Changed lines 224-225 from:
to:
./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/logic/sampleNet.txt [[<<]] -patterns -isettings inFiles/exts/exts/Patterns/settings.txt -n 5 -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt -calc myFirstCalcSimulation July 23, 2008, at 05:06 PM
by -
Changed line 224 from:
to:
July 23, 2008, at 05:05 PM
by -
Changed line 224 from:
to:
July 23, 2008, at 04:59 PM
by -
Added lines 117-119:
ChemChains Output
Depending on which mode ChemChains is run in, different output will be generated. In general, both the visual and calculation modes will create and save their output in a directory called 'CCOutput' that will be created (if it already doesn't exist) in a directory directly above the directory with ChemChains software. Refer to the Running Modes section on detailed description of output generated by ChemChains. Changed lines 150-151 from:
Although, ChemChains is a command-line program, it can output visual simulation results via its visual mode. To run ChemChains in visual mode, use the “-vis” flag. to:
Although, ChemChains is a command-line program, it can output visual simulation results via its visual mode. To run ChemChains in visual mode, use the “-vis” flag. To simulate the toy network using the simulation specification file both created in the previous sections run ChemChains as follows:
Note: Make sure that you have created at least one output node in the simulation specification file! Once the simulation finishes, a new file will be created in the ChemChains output folder, in a text file called “myFirstSimulation.txt”. Open the text file and we should see output similar to the Figure 3. ('*' denotes the active state of the node, while '.' represents the OFF state) Note: When running in the visual mode, ChemChains can run only one simulation at a time. To run multiple simulations in an automated fashion, run ChemChains in calculation mode. Changed lines 163-166 from:
to:
Calculation mode provide ChemChains users the ability to run thousands of automated simulations and subsequently use the generated data to analyze the dynamics of the simulated model. To run ChemChains in the calculation mode, use the '-calc' flag. Other flags available within the calculation mode are listed in Table 2.
You can run ChemChains (using our previously created network descriptor and simulation specification files) in the calculation mode in the most basic way (without the use of any extensions) as follows:
This command will simulate our toy network five times and will save all output in CCOutput/stats/myFirstCalcSimulation folder. The following table describes the structure and contents of the output folder.
Patterns Analysis
To perform patterns analysis, ChemChains must be initiated in the calculation mode (as described above), and using '-patterns' flag. When using patterns to perform analysis, two additional input files are required: i) the settings file and ii) a text file with a list of nodes that will undergo the pattern analysis. Settings File
The patterns settings file defines activity ranges that will form patterns. For your convenience, a sample settings files has already been created. You can find it and edit it in inFiles/exts/Patterns/settings.txt. The file contains two groups of settings: i) default settings that are applied on all nodes subjected to patterns analysis and ii) node specific settings that apply to only one node at a time. The syntax is as follows: Default settings: default:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2
The sample settings provided should have the following: default:0 9 0;10 29 1;30 100 2,
which means the activity of all nodes being analyzed will be categorized into three ranges: 0-9% (all nodes in this range will be coded “0”), 10-29% (all nodes in this range will be coded “1”), and 30-100% (all nodes in this range will be coded “2”) Node specific settings: NodeName:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2
List of nodes to be analyzed
An example of this file is also supplied with the latest ChemChains release, and is located in inFiles/exts/Patterns/sampleOutNodes.txt. In this file, each node to be analyzed with the patterns extension is entered on a new line. To run ChemChains using these input files, type in the following command:
July 22, 2008, at 10:19 PM
by -
Added lines 1-153:
Requirements
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms. Installation
Linux
Windows
Basic How-to
Introduction
This section will provide you with the most basic ways of simulating your first network model using ChemChains. ./ChemChains -h
Input Files
To simulate logical networks with ChemChains, two input files are required: the network descriptor and a simulation specification file. Network Descriptor
Network descriptor is a syntax-specific text file that stores the network, and logic for each node.  Figure 1: Sample network. As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows:
 Figure 2: Truth tables for nodes A, B, and C. These activation mechanisms can be expressed as individual truth tables (Figure 2). Once we have the network and truth tables established, we can create our network descriptor file using these tables. To create the file, the following syntax needs to be used: Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma)
Thus, the network descriptor for our toy network can look like: Bool:A:False:C:T
Bool:B:False:A,C:FF,FT,TT
Bool:C:False:B:T
Note that the declaration of each node is on a new line.
Once finished we can save the file as Simulation Specification file
ChemChains is a feature-rich logic network simulation software, which offers users many advanced simulation options. These options are specified by users in the specification file, the second of the required input files, that is loaded into the program before each simulation experiment. The following describes the various options contained within this file:
Syntax: For our example, we'll set the total time to 20 iterations and transient time to 10 as follows: RunTime:10:20
Syntax to declare an input (note that each input needs to be declared on a new line: Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below)
To declare input IN1, with True as the initial condition, fixed percentage activity on a periodic cycle set to 10%, and effective for the entire simulation length add the following line in the specification file: Input:IN1:True:F:F:1:10:20
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level, at time t, will be 10%+/-5.
To measure the current level of activity for each input during a simulation, ChemChains uses a sliding window (user pre-defined length) approach which allows the program to know whether the activity is within the desired percentage ON. In this example we'll set the sliding window to 5.
Syntax: Noise:5:5
The mutation parameter enables users to turn ON/OFF any node in the network to perform mutagenesis studies. When the node(s) is mutated, the corresponding truth table is ignored by the simulation engine and the activity of the node becomes fixed for the entire course of the simulation. For example, to turn off node A, you would add the following line to the specification file: Mutation:OFF:A
To mutate node A to be constitutively active, replace 'OFF' with 'ON'.
[Needs content]
[Needs content]
[Needs content]
Running Modes
The most recent version of ChemChains can be ran in three modes, one of which is a network construction mode (or File Conversion) and the remaining two are simulation modes. File Conversion Mode
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes. These truth tables need to be in the form of a tab-delimited text file. Each table must be in a separate file which should be named the same as the name of the node of interest. Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. In addition, we need to create another file with the list of nodes comprising our network. In this file, each node has two properties, nodeID and initial value, associated with it. Each node and its properties need to be on a new line. The mentioned node properties need to be tab-separated. The syntax is as follows: NodeName NodeID InitialValue
Sample node list for our network will look like: A 1 0
B 2 0
C 3 0
IN1 IN101 1
Note that if the initial value is False (i.e., 0 ) we can leave the third column empty.
Save the file as To create the Network Descriptor file from existing truth tables, run ChemChains with the following parameters: ./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet
Visual Mode
Although, ChemChains is a command-line program, it can output visual simulation results via its visual mode. To run ChemChains in visual mode, use the “-vis” flag. Calculation Mode
[Needs content] [Needs more content...] Advanced How-to
[Needs content] July 18, 2008, at 09:19 PM
by -
Deleted lines 0-144:
Requirements
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms. Installation
Linux
Windows
Basic How-to
Introduction
This section will provide you with the most basic ways of simulating your first network model using ChemChains. ./ChemChains -h
Input Files
To simulate logical networks with ChemChains, two input files are required: the network descriptor and a simulation specification file. Network Descriptor
Network descriptor is a syntax-specific text file that stores the network, and logic for each node.  Figure 1: Sample network. As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows:
 Figure 2: Truth tables for nodes A, B, and C. These activation mechanisms can be expressed as individual truth tables (Figure 2). Once we have the network and truth tables established, we can create our network descriptor file using these tables. To create the file, the following syntax needs to be used: Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma)
Thus, the network descriptor for our toy network can look like: Bool:A:False:C:T
Bool:B:False:A,C:FF,FT,TT
Bool:C:False:B:T
Note that the declaration of each node is on a new line.
Once finished we can save the file as Simulation Specification file
ChemChains is a feature-rich logic network simulation software, which offers users many advanced simulation options. These options are specified by users in the specification file, the second of the required input files, that is loaded into the program before each simulation experiment. The following describes the various options contained within this file:
Syntax: For our example, we'll set the total time to 20 iterations and transient time to 10 as follows: RunTime:10:20
Syntax to declare an input (note that each input needs to be declared on a new line: Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below)
To declare input IN1, with True as the initial condition, fixed percentage activity on a periodic cycle set to 10%, and effective for the entire simulation length add the following line in the specification file: Input:IN1:True:F:F:1:10:20
[Needs content]
[Needs content]
[Needs content]
[Needs content]
[Needs content]
Simulation Modes
The most recent version of ChemChains can be ran in three modes, one of which is a network construction mode (or File Conversion) and the remaining two are simulation modes. File Conversion Mode
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes. These truth tables need to be in the form of a tab-delimited text file. Each table must be in a separate file which should be named the same as the name of the node of interest. Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. In addition, we need to create another file with the list of nodes comprising our network. In this file, each node has two properties, nodeID and initial value, associated with it. Each node and its properties need to be on a new line. The mentioned node properties need to be tab-separated. The syntax is as follows: NodeName NodeID InitialValue
Sample node list for our network will look like: A 1 0
B 2 0
C 3 0
IN1 IN101 1
Note that if the initial value is False (i.e., 0 ) we can leave the third column empty.
Save the file as To create the Network Descriptor file from existing truth tables, run ChemChains with the following parameters: ./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet
Visual Mode
[Needs content] Calculation Mode
[Needs content] [Needs more content...] Advanced How-to
[Needs content] July 18, 2008, at 09:18 PM
by -
Added line 98:
Deleted lines 99-100:
[Needs content]
July 18, 2008, at 09:16 PM
by -
Changed lines 84-85 from:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage
to:
Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage (explanation of each option below)
Changed lines 96-97 from:
Input:name:True:F:F:1:10:20
to:
Input:IN1:True:F:F:1:10:20
July 18, 2008, at 08:12 PM
by -
Changed lines 82-83 from:
Syntax to declare an input (note that each input needs to be declared on a new line) to:
Syntax to declare an input (note that each input needs to be declared on a new line: Added lines 85-89:
To declare input IN1, with True as the initial condition, fixed percentage activity on a periodic cycle set to 10%, and effective for the entire simulation length add the following line in the specification file: Input:name:True:F:F:1:10:20
July 18, 2008, at 08:06 PM
by -
Changed lines 73-83 from:
to:
Syntax: For our example, we'll set the total time to 20 iterations and transient time to 10 as follows: RunTime:10:20
Syntax to declare an input (note that each input needs to be declared on a new line) Input:name:initial/default value:random(R)/fixed(F):noise(N)/fixed/(F):time to be introduced:dose (in percentage):duration of the dosage
Deleted lines 84-85:
[Needs content]
July 17, 2008, at 05:07 PM
by -
Added lines 90-91:
The most recent version of ChemChains can be ran in three modes, one of which is a network construction mode (or File Conversion) and the remaining two are simulation modes. Added lines 117-121:
Visual Mode
[Needs content] Calculation Mode
[Needs content] July 17, 2008, at 04:47 PM
by -
Changed line 90 from:
File Conversion
to:
File Conversion Mode
Changed lines 95-96 from:
Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format. to:
Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. Changed lines 109-110 from:
to:
Save the file as To create the Network Descriptor file from existing truth tables, run ChemChains with the following parameters: ./ChemChains -TT2ND -iNodeList inFiles/exts/FileConverter/sampleNodeList.txt -iTables inFiles/exts/FileConverter/sampleNet
July 17, 2008, at 04:37 PM
by -
Added lines 100-102:
The syntax is as follows: NodeName NodeID InitialValue
Changed lines 108-110 from:
to:
Note that if the initial value is False (i.e., 0 ) we can leave the third column empty.
July 17, 2008, at 04:34 PM
by -
Added lines 97-105:
In addition, we need to create another file with the list of nodes comprising our network. In this file, each node has two properties, nodeID and initial value, associated with it. Each node and its properties need to be on a new line. The mentioned node properties need to be tab-separated. Sample node list for our network will look like: A 1 0
B 2 0
C 3 0
IN1 IN101 1
July 17, 2008, at 04:28 PM
by -
Changed lines 90-92 from:
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes.
to:
File Conversion
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes. July 17, 2008, at 04:27 PM
by -
Added lines 90-96:
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes.
These truth tables need to be in the form of a tab-delimited text file. Each table must be in a separate file which should be named the same as the name of the node of interest. Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format. July 17, 2008, at 04:16 PM
by -
Changed lines 17-18 from:
To run ChemChains in Windows, a Linux-like environment is necessary. For example, you can download and install the freely available cygwin from here. to:
To run ChemChains in Windows, a Linux-like environment is necessary. For example, you can download and install the freely available cygwin from here. July 16, 2008, at 10:27 PM
by -
Changed line 75 from:
to:
[Needs content]
Added line 77:
[Needs content]
Added line 79:
[Needs content]
Added line 81:
[Needs content]
Added line 83:
[Needs content]
Changed lines 85-86 from:
to:
[Needs content]
July 16, 2008, at 10:27 PM
by -
Changed line 73 from:
[Needs content] to:
[Needs content]
July 16, 2008, at 10:27 PM
by -
Added line 73:
[Needs content] Added line 75:
July 16, 2008, at 10:26 PM
by -
Changed lines 65-69 from:
Once finished we can save the file as
to:
Once finished we can save the file as Changed lines 68-79 from:
to:
ChemChains is a feature-rich logic network simulation software, which offers users many advanced simulation options. These options are specified by users in the specification file, the second of the required input files, that is loaded into the program before each simulation experiment. The following describes the various options contained within this file:
July 16, 2008, at 10:03 PM
by -
Changed lines 58-59 from:
Thus, our toy network would look like: to:
Thus, the network descriptor for our toy network can look like: Changed lines 64-66 from:
to:
Note that the declaration of each node is on a new line.
Once finished we can save the file as July 16, 2008, at 10:01 PM
by -
Changed lines 58-66 from:
to:
Thus, our toy network would look like: Bool:A:False:C:T
Bool:B:False:A,C:FF,FT,TT
Bool:C:False:B:T
July 16, 2008, at 09:57 PM
by -
Changed lines 54-58 from:
to:
Once we have the network and truth tables established, we can create our network descriptor file using these tables. To create the file, the following syntax needs to be used: Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma)
July 16, 2008, at 05:07 PM
by -
Changed lines 42-43 from:
 Figure 1. to:
 Figure 1: Sample network. Changed lines 50-51 from:
 Figure 2. to:
 Figure 2: Truth tables for nodes A, B, and C. July 16, 2008, at 05:04 PM
by -
Changed line 47 from:
to:
July 16, 2008, at 04:55 PM
by -
Changed line 47 from:
to:
July 16, 2008, at 04:54 PM
by -
Changed lines 52-54 from:
These activation mechanisms can be expressed as a truth tables (Figure 2). to:
These activation mechanisms can be expressed as individual truth tables (Figure 2). July 16, 2008, at 04:53 PM
by -
Changed lines 50-51 from:
Attach:toynetwork_TT.jpg Δ | Figure 2. to:
 Figure 2. July 16, 2008, at 04:53 PM
by -
Changed lines 50-51 from:
Attach:toynetwork_TT.jpeg Δ | Figure 2. to:
Attach:toynetwork_TT.jpg Δ | Figure 2. July 16, 2008, at 04:53 PM
by -
Changed lines 50-52 from:
This mechanism can be expressed as a truth table. to:
Attach:toynetwork_TT.jpeg Δ | Figure 2. These activation mechanisms can be expressed as a truth tables (Figure 2). July 16, 2008, at 03:43 PM
by -
Added line 45:
July 16, 2008, at 03:43 PM
by -
Changed line 44 from:
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. The activation mechanisms for the nodes of this network are as follows: to:
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. Let's assume the activation mechanisms for the nodes of this network as follows: July 16, 2008, at 03:42 PM
by -
Added lines 42-43:
 Figure 1. Changed line 51 from:
 Figure 1. to:
July 16, 2008, at 03:41 PM
by -
Changed lines 42-43 from:
As an example, let's assume the following toy network consisting of nodes A, B, and C. to:
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, and C, where green line (with an arrow) denotes positive regulation (i.e., activation) and red line (with a stop line) denotes negative regulation. Because this is a boolean network, each node has a an activation mechanism associated with it. The activation mechanisms for the nodes of this network are as follows:
This mechanism can be expressed as a truth table. July 16, 2008, at 03:35 PM
by -
Changed lines 44-45 from:
 Figure 1. to:
 Figure 1.
July 16, 2008, at 03:34 PM
by -
Changed lines 44-45 from:
 Figure 1. to:
 Figure 1. July 16, 2008, at 03:33 PM
by -
Changed lines 44-45 from:
 Figure 1. to:
 Figure 1. July 16, 2008, at 03:33 PM
by -
Changed lines 44-46 from:
%lfloat text-align=center margin-top=5px margin-right=25px
margin-bottom=5px margin-left=25px% to:
 Figure 1. July 16, 2008, at 03:32 PM
by -
Changed lines 44-45 from:
 to:
%lfloat text-align=center margin-top=5px margin-right=25px
margin-bottom=5px margin-left=25px% July 16, 2008, at 03:26 PM
by -
Changed lines 44-46 from:
 toynet.jpeg"Sample toy network" to:
 July 16, 2008, at 02:58 PM
by -
Changed lines 44-46 from:
to:
 toynet.jpeg"Sample toy network" July 16, 2008, at 02:26 PM
by -
Changed lines 44-46 from:
 to:
 July 16, 2008, at 02:25 PM
by -
Changed lines 44-46 from:
 to:
 July 16, 2008, at 02:08 PM
by -
Changed lines 45-48 from:
http://example.com| to:
July 16, 2008, at 02:07 PM
by -
Changed line 45 from:
 to:
http://example.com| Added line 48:
July 16, 2008, at 02:04 PM
by -
Changed lines 46-47 from:
to:
http://pmichaud.com/img/misc/pc.jpg | [- [[ (Wikipedia:Paper_clips | Paper clips ]] are fun to work with. -] July 16, 2008, at 02:03 PM
by -
Changed lines 45-46 from:
to:
 July 16, 2008, at 02:03 PM
by -
Changed lines 45-46 from:
to:
July 16, 2008, at 02:00 PM
by -
Changed lines 44-45 from:
to:
 July 16, 2008, at 01:57 PM
by -
Changed lines 44-45 from:
to:
July 16, 2008, at 01:56 PM
by -
Changed lines 44-45 from:
 to:
July 16, 2008, at 01:56 PM
by -
Changed lines 44-45 from:
to:
 July 16, 2008, at 01:55 PM
by -
Changed lines 44-45 from:
 to:
July 16, 2008, at 01:43 PM
by -
Added line 45:
July 16, 2008, at 01:35 PM
by -
Changed line 44 from:
to:
 July 16, 2008, at 01:05 PM
by -
Changed lines 40-42 from:
Network descriptor is a syntax-specific text file that stores the network and logic for each node. to:
Network descriptor is a syntax-specific text file that stores the network, and logic for each node. As an example, let's assume the following toy network consisting of nodes A, B, and C. July 16, 2008, at 01:03 PM
by -
Changed lines 40-42 from:
to:
Network descriptor is a syntax-specific text file that stores the network and logic for each node. July 16, 2008, at 12:59 PM
by -
Changed lines 39-42 from:
Network Descriptor
Simulation Specification file
to:
Network Descriptor
Simulation Specification file
July 16, 2008, at 12:58 PM
by -
Changed line 29 from:
to:
Changed line 36 from:
to:
Changed lines 43-44 from:
to:
July 16, 2008, at 12:57 PM
by -
Changed lines 37-42 from:
to:
To simulate logical networks with ChemChains, two input files are required: the network descriptor and a simulation specification file. Network Descriptor
Simulation Specification file
July 16, 2008, at 12:28 AM
by -
Changed line 1 from:
to:
July 16, 2008, at 12:27 AM
by -
Added lines 8-9:
Changed lines 35-36 from:
to:
July 16, 2008, at 12:24 AM
by -
Changed lines 28-29 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag.\\ to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. July 16, 2008, at 12:24 AM
by -
Changed line 28 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag.\\ July 16, 2008, at 12:24 AM
by -
Changed lines 28-32 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag.
to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag. ./ChemChains -h
July 16, 2008, at 12:23 AM
by -
Changed lines 29-34 from:
to:
July 16, 2008, at 12:22 AM
by -
Changed line 30 from:
./ChemChains -h
to:
./ChemChains -h July 16, 2008, at 12:22 AM
by -
Changed lines 28-31 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. To see all available options and command-line parameters run ChemChains with the -h flag.
./ChemChains -h
July 16, 2008, at 12:16 AM
by -
Changed lines 28-31 from:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. July 16, 2008, at 12:16 AM
by -
Changed lines 28-31 from:
To run ChemChains, users must provide several input files (described below). Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. to:
To run ChemChains, users must provide several input files. Once simulations and/or analysis are initiated, ChemChains will create several types of output file formats. Both the input files, as well as the desired output format and the output files must be specified by users in the form of command-line parameters provided to ChemChains at the time of program initiation. July 16, 2008, at 12:15 AM
by -
Changed line 26 from:
This section will provide you with the most basic ways of simulating your first network model using ChemChains.\\ to:
This section will provide you with the most basic ways of simulating your first network model using ChemChains.\\ July 16, 2008, at 12:14 AM
by -
Changed line 12 from:
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms. \\ to:
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms.\\ July 16, 2008, at 12:14 AM
by -
Changed lines 26-27 from:
This section will provide you with the most basic ways of simulating your first network model using ChemChains.\\ to:
This section will provide you with the most basic ways of simulating your first network model using ChemChains.\\ \\ July 16, 2008, at 12:14 AM
by -
Changed line 12 from:
ChemChains is a platform independent software written in C++, that has been tested on Linux and Windows platforms. \\ to:
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms. \\ Changed lines 26-32 from:
[Needs content] to:
This section will provide you with the most basic ways of simulating your first network model using ChemChains.\\ [Needs more content...] July 16, 2008, at 12:08 AM
by -
Changed line 12 from:
ChemChains is a platform independent software that has been tested on Linux and Windows platforms. \\ to:
ChemChains is a platform independent software written in C++, that has been tested on Linux and Windows platforms. \\ July 16, 2008, at 12:04 AM
by -
Deleted line 24:
Changed lines 27-28 from:
to:
July 16, 2008, at 12:04 AM
by -
Changed line 27 from:
to:
[Needs content] July 16, 2008, at 12:03 AM
by -
Changed lines 17-25 from:
to:
Linux
Windows
July 15, 2008, at 11:58 PM
by -
Changed line 13 from:
to:
\\ July 15, 2008, at 11:58 PM
by -
Changed lines 14-15 from:
To run ChemChains in Windows, download cygwin to:
To run ChemChains in Windows, a Linux-like environment is necessary. For example, you can download and install the freely available cygwin from here. July 15, 2008, at 11:56 PM
by -
Changed lines 12-21 from:
alkhdflakshdf to:
ChemChains is a platform independent software that has been tested on Linux and Windows platforms. July 15, 2008, at 11:23 PM
by -
Added line 21:
July 15, 2008, at 11:23 PM
by -
Changed lines 12-20 from:
to:
alkhdflakshdf July 15, 2008, at 11:22 PM
by -
Changed lines 6-8 from:
to:
Added lines 12-17:
July 15, 2008, at 11:21 PM
by -
Changed lines 9-10 from:
to:
July 15, 2008, at 11:21 PM
by -
Changed lines 9-10 from:
to:
July 15, 2008, at 11:21 PM
by -
Changed lines 9-10 from:
to:
July 15, 2008, at 11:13 PM
by -
Changed lines 6-8 from:
to:
July 15, 2008, at 11:12 PM
by -
Changed line 1 from:
to:
Changed line 5 from:
to:
July 15, 2008, at 11:08 PM
by -
Changed lines 5-6 from:
to:
Deleted lines 10-14:
July 15, 2008, at 11:07 PM
by -
Added lines 1-8:
July 15, 2008, at 11:04 PM
by -
Changed lines 4-5 from:
to:
July 15, 2008, at 11:02 PM
by -
Changed lines 1-4 from:
to be written... to:
July 15, 2008, at 11:00 PM
by -
Added line 1:
to be written... |