|
Table of Contents
1. Requirements
2. Installation 2.1. Download 2.2. Linux 2.3. Windows 2.3.1. Installing Cygwin 2.3.2. Running Cygwin & Setting up ChemChains 3. Basic How-to 3.1. Introduction 3.2. Input Files 3.2.1. Network Descriptor 3.2.1.1. Flag to Specify the Network Descriptor File 3.2.2. Simulation Specification file 3.2.2.1. Flag to Specify the Simulation Specification File 3.3. ChemChains Output 3.4. Running Modes 3.4.1. File Conversion Mode 3.4.2. Visual Mode 3.4.3. Calculation Mode 3.4.3.1. Patterns Analysis 3.4.3.2. Random Initial Conditions 1. Requirements
ChemChains is a platform independent command-line software written in C++, that has been tested on Linux and Windows platforms. 2.1. Download
2.2. Linux
2.3.1. Installing Cygwin
2.3.2. Running Cygwin & Setting up ChemChains
Congratulations!! ChemChains is now installed and ready for your first simulation! Some basic Unix commands
3.1. Introduction
This section will provide you with the most basic ways of simulating your first network model using ChemChains. ./ChemChains -h
3.2. Input Files
To simulate logical networks with ChemChains, two input files are required: the network descriptor file and simulation specification file (additional input files may be required for individual running modes. 3.2.1. Network Descriptor
Network descriptor is a syntax-specific text file that stores the network, and logic for each node.
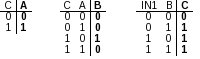
As an example, let's assume a toy network (Figure 1) consisting of nodes A, B, C, and external input IN1, where green line (with an arrow) denotes positive regulation (e.g., activation) and red line (with a stop line) denotes negative regulation (e.g., inhibition). Because we're dealing with a boolean network, each node has an activation mechanism (represented by truth tables) associated with it. As an example, the activation mechanisms for the nodes of this network are as follows:
These activation mechanisms can be expressed as individual truth tables (Figure 2). Once the network and truth tables are established, they can be used to create the network descriptor file. Use the following syntax to create the file: Bool:name:initial state[True|False]:input1,input2..inputN:list of states of input nodes for non-initial state(separated by comma) Thus, the network descriptor for our toy network can look like: Note: The declaration of each node is on a new line. Bool:A:False:C:T
Bool:B:True:C,A:FF,FT,TT
Bool:C:True:IN1,B:FF
Once finished, save the file as 3.2.1.1. Flag to Specify the Network Descriptor File
./ChemChains -ilogic [network descriptor filename]
3.2.2. Simulation Specification file
ChemChains is a feature-rich logic network simulation software, which offers users many advanced simulation options. These options are specified by users in the specification file, the second of the required input files, that is loaded into the program before each simulation experiment. (The following example of a simple simulation specification file is included with the current ChemChains release and can be found in "inFiles/CChain/specs/sampleSpecs.txt" file in the ChemChains directory) The following describes the various options contained within this file:
Syntax: For our example, we'll set the total time to 100 iterations and transient time to 70 as follows: RunTime:70:100
Syntax to declare an input: Input:name:initial/default value:random(R)/fixed(F): noise(N)/fixed/(F):time to be introduced:dose (in percentage): duration of the dosage Note: Each input needs to be declared on a new line.
To declare input IN1, with True as the initial condition, fixed percentage activity on a periodic cycle set to 10%, and effective for the entire simulation length add the following line in the specification file: Input:IN1:True:F:F:1:10:99
Advanced Example: For each simulation, to set IN1 between 0-10% for the first 30 iterations, 11-50% for iterations 31-70, and 51-100% for the remaining simulation time, add the following:
Input:IN1:True:R:F:1:0-10:29,30:11-50:39,71:51-100:29
The noise variable allows the input activity level to randomly bounce around the defined value within a specified range. For example, if the activity level for IN1 is set to 10% with a noise range of 5, the actual activity level at time t will be 10+/-5%.
To measure the current level of activity for each input during a simulation, ChemChains uses a sliding window (user pre-defined length) approach which allows the program to know whether the activity is within the desired percentage ON. In this example we'll set the sliding window to 5.
Syntax: Noise:5:5
The mutation parameter enables users to turn ON/OFF any node in the network to perform mutagenesis studies. When the node(s) is mutated, the corresponding truth table is ignored by the simulation engine and the activity of the node becomes fixed for the entire course of the simulation. For example, to turn off node A, you would add the following line to the specification file: Mutation:OFF:A
To mutate node A to be constitutively active, replace 'OFF' with 'ON'. Note: The name of the delay, sustain, and output nodes has to differ from the names of their inputs.
To create a delay node, use the following syntax: Delay:name:initial value:parent node:iterations to wait
To create a sustain node, use the following syntax: Sustain:name:initial value:parent node:duration
Output nodes are nodes that will be printed out via ChemChains' visual mode. To create a new output node, use the following syntax: Output:output name:initial value:node name
3.2.2.1. Flag to Specify the Simulation Specification File
./ChemChains -ispecs [simulation specification filename]
3.3. ChemChains Output
Depending on which mode ChemChains is run in, different output will be generated. In general, both the visual and calculation modes will create and save their output in a directory called 'CCOutput' that will be created (if it already doesn't exist) in a directory directly above the directory with ChemChains software. Refer to the Running Modes section on detailed description of output generated by ChemChains. 3.4. Running Modes
The most recent version of ChemChains can be ran in three modes, one of which is a network construction mode (or File Conversion) and the remaining two are simulation modes. 3.4.1. File Conversion Mode
As mentioned above, to run ChemChains, a file with the network description is required. This file can be created manually as described in the previous section, or automatically from a provided set of truth tables for individual nodes. These truth tables need to be in the form of a tab-delimited text file. Each table must be in a separate file which should be named the same as the name of the node of interest. Thus, for the toy network (Figure 1) with truth tables as shown in Figure 2, create a new directory called "sampleNet" in "inFiles/exts/FileConverter", and three text files (A.csv, B.csv, and C.csv) with the truth tables in tab-delimited format and save them in the "inFiles/exts/FileConverter/sampleNet" directory. In addition, we need to create another file with the list of nodes comprising our network. In this file, each node has two properties, nodeID and initial value, associated with it. Each node and its properties need to be on a new line. The mentioned node properties need to be tab-separated. The syntax is as follows: NodeName NodeID InitialValue
Note: If the initial value is False (i.e., 0 ) we can leave the third column empty. Sample node list for our network will look like: A 1
B 2 1
C 3 1
IN1 IN4
Save the file as To create the Network Descriptor file from existing truth tables, run ChemChains with the following parameters: ./ChemChains -TT2ND -inodelist inFiles/exts/FileConverter/sampleNodeList.txt -itables inFiles/exts/FileConverter/sampleNet -o outputFileName.txt 3.4.2. Visual Mode
Although, ChemChains is a command-line program, it can output visual simulation results via its visual mode. To run ChemChains in visual mode, use the “-vis” flag. To simulate the toy network using the simulation specification file both created in the previous sections run ChemChains as follows: ./ChemChains -ilogic inFiles/exts/CChain/logic/sampleNet.txt -ispecs inFiles/exts/CChain/specs/sampleSpecs.txt -vis myFirstSimulation.txt Note: Make sure that you have created at least one output node in the simulation specification file! Once the simulation finishes, a new file will be created in the ChemChains output folder, in a text file called “myFirstSimulation.txt”. Open the text file and we should see output similar to the Figure 3. ('*' denotes the active state of the node, while '.' represents the OFF state) Note: When running in the visual mode, ChemChains can run only one simulation at a time. To run multiple simulations in an automated fashion, run ChemChains in calculation mode. 3.4.3. Calculation Mode
Calculation mode provide ChemChains users the ability to run thousands of automated simulations and subsequently use the generated data to analyze the dynamics of the simulated model. To run ChemChains in the calculation mode, use the '-calc' flag. You can run ChemChains (using our previously created network descriptor and simulation specification files) in the calculation mode in the most basic way (without the use of any extensions) as follows: ./ChemChains -ilogic inFiles/CChain/logic/sampleNet.txt -ispecs inFiles/CChain/specs/sampleSpecs.txt -n 5 -calc myFirstCalcSimulation This command will simulate our toy network five times and will save all output in CCOutput/stats/myFirstCalcSimulation folder. The following table describes the structure and contents of the output folder.
3.4.3.1. Patterns Analysis
To perform patterns analysis, ChemChains must be initiated in the calculation mode (as described above), and using '-patterns' flag. When using patterns to perform analysis, two additional input files are required: i) the settings file and ii) a text file with a list of nodes that will undergo the pattern analysis.
The patterns settings file defines activity ranges that will form patterns. For your convenience, a sample settings files has already been created. You can find it and edit it in inFiles/exts/Patterns/settings.txt. The file contains two groups of settings: i) default settings that are applied on all nodes subjected to patterns analysis and ii) node specific settings that apply to only one node at a time. The syntax is as follows: Default settings: default:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2 The sample settings provided should have the following: default:0 9 0;10 29 1;30 100 2,
which means the activity of all nodes being analyzed will be categorized into three ranges: 0-9% (all nodes in this range will be coded “0”), 10-29% (all nodes in this range will be coded “1”), and 30-100% (all nodes in this range will be coded “2”) Node specific settings: NodeName:lower_limit1 higher_limit1 code1, lower_limit2 higher_limit2 code2
An example of this file is also supplied with the latest ChemChains release, and is located in inFiles/exts/Patterns/sampleOutNodes.txt. In this file, each node to be analyzed with the patterns extension is entered on a new line. To run ChemChains using these input files, type in the following command: ./ChemChains -ilogic inFiles/CChain/logic/sampleNet.txt -ispecs inFiles/CChain/specs/sampleSpecs.txt -patterns -isettings inFiles/exts/Patterns/settings.txt -inodes nFiles/exts/exts/Patterns/sampleOutNodes.txt -n 5 -calc myFirstCalcSimulation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||