Metannogen Tutorial
This tutorial demonstrates how Metannogen is used to annotate reactions in published networks or to reconstruct metabolic networks. Start Metannogen by clicking The initial download of KEGG may take several minutes.
A window to select the dataset source will appear.
Three networks are loaded: KEGG, EHMN and Reccon1.
Then choose the Demo dataset server by pressing "For testing use demo dataset".
This button inserts the URL of the demo repository into the text field for the central dataset server.
The "Password" field can remain empty.
Previously entered URLs are commented out by a "#"-sign.
They can be reactivated by removing the "#"-sign.
Now press the "Go"-button to begin the Metannogen session.
Please note that the "Go"-button remains inactive until all
downloads have been completed.
The initial download of KEGG may take several minutes.
A window to select the dataset source will appear.
Three networks are loaded: KEGG, EHMN and Reccon1.
Then choose the Demo dataset server by pressing "For testing use demo dataset".
This button inserts the URL of the demo repository into the text field for the central dataset server.
The "Password" field can remain empty.
Previously entered URLs are commented out by a "#"-sign.
They can be reactivated by removing the "#"-sign.
Now press the "Go"-button to begin the Metannogen session.
Please note that the "Go"-button remains inactive until all
downloads have been completed.
General Appearance
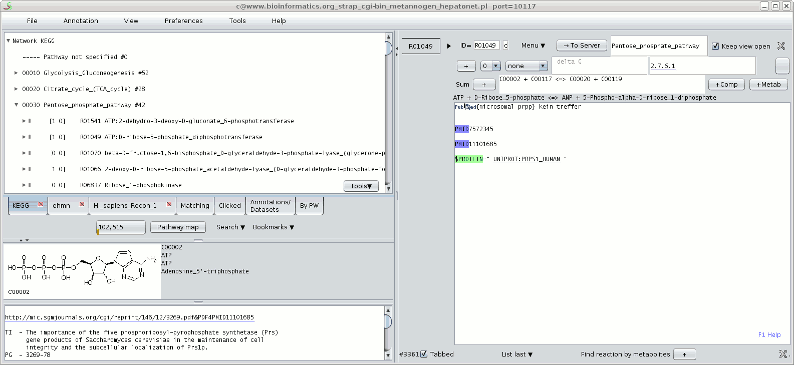
After pressing the "Go"-button Metannogen comes along with a GUI (see Figure). The GUI shows a menu-bar at the top. The left panel and a right panel is divider by a divider which can be dragged with the mouse.The left panels
 |
- Tab "KEGG", tab "RECON1" and tab "EHMN": The first tabs contain immutable reaction networks. Other networks can be included (Menu-bar>File>Import ... ). Look at the network items by opening tree nodes. In the EHMN network no pathways are assigned to reactions and therefore the reactions appear as a flat list.
- Tab "Clicked":
This tab contains objects that are clicked in text elements.
Please open a notepad (Menu-bar>Tools) and type
NETWORK#R00014
Click on this reference and the Kegg reaction R00014 appears in the tree "Clicked" of objects.
Note: You can toggle "Always floating on top of other frames" for this notepad by typing Ctrl-T. The state is indicated in the Application frame. This applies to all windows of Metannogen. - Tab "Matching": This tab lists items that match a string pattern. Open the search-dialog by pressing one of the "Search-Data" tool buttons. Perform a search for "glucose". All reactions, compounds and datasets that contain the text pattern "glucose" will appear in this tab.
- Tab "Datasets": The tab "Datasets" provides a list of all loaded datasets of the user model. Please create a new dataset and find the new dataset in the list of datasets as described below.
- Tab "By Pathway": Here the Datasets are sorted according to the Pathway that was typed into the "Pathway" text fields of the datasets. The list is updated when the mouse enters the panel. Please type something into the "Pathway" text fields of a dataset and watch this tree.
C00004into the notepad. Move the mouse over C00004. The chemical structure should appear in the middle panel. The left lower panel contains textural information of individual reactions, enzymes or compounds.
Automatical download of abstracts and sequence files: Type into the notepad
UNIPROT:hslv_ecoli PMID16697045 ENSG00000088826Move the mouse pointer over each single text token. Wait a few seconds to give it sufficient time for downloading. Then move the mouse pointer again over these tokens. The text information will appear.
The right panel
In the right panel, datasets are edited. The user can modify the information using text-fields, choice menus and toggle buttons.Annotate reactions
Open the KEGG network tree (first tab in the upper left panel) and open the of an enzymatic reaction, i.e. expand a pathway node and chose one of the reactions. Then, click the menu item " New dataset". A new dataset for this enzymatic reaction is created.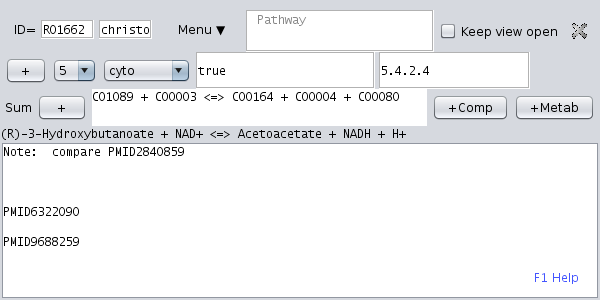 |
Persistance of data
Finally press the upload-tool button "-> To Server".The data is saved permanently: You can close the dataset and re-open it by clicking the respective reaction node or the node in the tab "Datasets" or the tab "By pathway". Finally, close the Metannogen and restart metannogen and find the dataset.
Menubar
Frequently used menu items can be undocked. This is useful for frequently used menu-items. Open the Dataset menu. Then drag the menu-item "Create Dataset from scratch" to the Desktop.Cross references
Dataset and Network references
Type references to the Kegg network into the multi-line text-field or into a notepad (Menu-bar>Tools). Examples:
NETWORK#R00004
KEGG#R00004
Click these with the left mouse button. Try the context menu by right click.
Try the context menu for different metabolites and reactions.
Try the prefix "DATASET#" such as "DATASET#R00004". If a dataset with the respective identifier exist, it would be opened upon mouse click.
Named text blocks
Type a named text block as the following into the multi-line text field of one dataset:
BEGIN#GlucoseTransport
There are glut1, glut2 ....
END#GlucoseTransport
Type a reference to this text block into another dataset. It might be embedded in a paragraph of free text.
#GlucoseTransport
or
INCLUDE#GlucoseTransport
Now click the reference or the BEGIN tag and watch.
External references
References may point to external data sources. Usually, external references have a colon or curly brackets and are underlined in blue. If they are clicked, the respective site is opened in the browser. Both HTTP protocols, POST and GET, are supported by metannogen. Examples with GET:
PUBMED{ glucose[TI] AND transport }
UNIPROT{ glucose }
GOOGLE{ glucose }
WIKI:glucose
METACYC{ glucose }
ENSG{ glucose }
Examples with POST
CITEXPLORE{ glucose }
ISI{ glucose } not yet working
TCDB{ glucose }
Examples with STRAP:
Aligning sequences: ALIGN{ SWISS:Q00612 SWISS:G6PD_RAT }
Predicting sub-cellular locations: LOC{ SWISS:G6PD_RAT }
Searching for similar sequences: BLAST{ SWISS:G6PD_RAT }
These references can be configured by the user: Menu-bar>Options>Web-Resources.
Literature Management
Abstract
Pubmed and UNIPROT entries have specific identifiers. Type the following into the multi-line comment text of a dataset:
PMID17408512
UNIPROT:P08559
UNIPROT:P06169
Now move the mouse over the identifiers and wait a few seconds for the abstract and protein files to be downloaded.
Now move the mouse again over these identifiers.
This time the abstract and protein file text is displayed in the left bottom panel.
If no abstract is visible, it might be that the text pane is squeezed to size zero and the divider bars must be dragged.
Full text
Right click the PMID17408512 reference and specify the download directory. Typical locations are /tmp/, $HOME/Desktop/ and $HOME/Downloads. File path completion can be used with the tabulator key. If you have problems then seek help from your system administrator.Manual PDF-file association: Now associate a PDF-document with the Pubmed reference PMID17408512. Open the respective dialog using the context menu (Right click). This dialog has a section for eacho of the three chronological steps:
- Section Preparation: Here you need to specify potential locations, where the browser might save downloaded PDF-files. It is likely, that the default settings are appropriate.
- Section Downloading: Here see a button with a web-browser icon which opens the Pubmed abstract. Visit the publishers site and download the PDF. If the PDF is shown within the browser, you need to save it. Anyway, it is recommended to change the browser settings such that PDFs are displayed in an external PDF viewer rather than in a browser tab. This can be done by changing the associations of the file type PDF in the settings of the browser in section "Applications".
- Section Downloading: The button "Associate" need to be pressed after downloading. Metannogen will find the PDF in the above directories that has been saved most recently. This should be your file. After successful association, the pmid-link turns red.
The capability of PDF viewers to highlight text is still limited. Therefore it might be useful to see the full text in Metannogen. Hold the alt-key and move the mouse slowly over the link. The full text is shown in Metannogen. Figures are not included. For some PDF files the text cannot be extracted and the text pane remains empty.
Automated PDF-file assignment: In about one third of all cases Metannogen may locate the PDF document automatically. The suggested links to the PDF full text appear above the abstract text and can be directly clicked.
Highlighting
To identify relevant text phrases in the publication abstracts and full-texts, certain keywords can be highlighted. A list of words to be highlighted can be specified for the entire project: "Menu-bar>Options>Search patterns ...".Add words and watch them being highlighted in the viewed abstracts.
Search patterns can also be selected for specific datasets using the variable "$HL" standing for (H)igh(L)ight. Type something like the following into the multi-line text field. Then move the mouse over the abstract text and watch:
$HL="glucose fructose "
Add web colors:
$HL=" #FF1130 glucose #00FF00 fructose "
Graphical Kegg Maps
The Kegg Maps for pathways can be opened with the context menu (Right click a reaction, pathway or EC-class). Check the following features:- Datasets with green and red traffic-lights are highlighted in the pathway map.
- Metabolites for which a transporter dataset is defined are highlighted.
- Reactions and metabolites have a context menu
- Objects may be marked by marching ants.
- Connections with other pathways are often shown. Clicking these pathway boxes opens the respective pathway view.
Input tools
Word expansion
When a user types the beginning of a long word and hits the tabulator key, Metannogen tries to complete the word. This is supported for all metabolite and reaction names in the loaded networks (Kegg, Palsson). Due to ambiguities, it may be completed to a word that is not the desired one. In this case the user can hit the tabulator key again until the desired word appears. Word completion is helpful for long complicated names and avoids mistyping of words.Balloon-Text
Move the mouse slowly over a metabolite identifier in the equation text field. A balloon text should appear telling the name of the metabolite. This works in all text fields of Metannogen and in the graphical pathway maps.Context-menu
Locate the mouse pointer on a metabolite or reaction identifier and click with the right mouse button. A Context menu should appear for the object under the mouse. Use this mechanism to locate the object in the graphical tree.Working in a Team of Curators
When several people are working on the same network, Metannogen needs to take care that data is not lost when two users are accessing and modifying the same dataset. The curator who uploads the dataset more recently could overwrite the changes of another curator who has changed the dataset at an earlier time. To avoid this, Metannogen warns users who edit a dataset, that has been just saved by another curator.Simulate this scenario by opening two metannogen sessions. Open the same dataset in both sessions and upload the dataset to the server in one session. Then try to modify the same dataset in the other Metannogen instance. Watch the warning message that appears.
Reaction Attributes
The multi-line text field contains free text which does not to be structured in a certain way. Usually, it is not possible to include information from this text-field in the exported model file. However, content that is captured in form of a variable will be available for further data processing. Add a simple variable declaration in the multi-line text field. If you know programming languages like or you will be familiar with the syntax.
$THE_FUNCTION=break-down
You can use any variable name starting with a letter and consisting of letters, digits and underscore.
It is recommended to register variable names in Menu-bar>Customize>Variables.
The advantage is that registered variables are color-highlighted.
This allows one to notice when variables are mistyped.
The file http://www.bioinformatics.org/strap/metannogen/data/annotationFormats.txt defines attributes that are inserted in rdf (Miriam) format into the resulting SBML file.
Using an input mask
Sometimes it is desired to record free-text data in each dataset in a standardized manner. For this purpose, input forms loaded with the command line option "-datasetForms" can be inserted into the multi-line text-field (menu-bar>datasets). The idea is to store information in a structured way that allows capturing of the information and its usage for the file export functions of Metannogen. Chose the first input mask from the "Forms"-menu. The form contains a number of empty variable declarations that can be filled by the user. The information is typed between the quotes. Simple variable-value assignments take the equals-sign whereas the plus-equals operator is used to append text content.
$SUPPORTING_EVI+=""
$SUPPORTING_EVI+=""
$SUPPORTING_EVI@cyto+=""
$SUPPORTING_EVI@mito+=""
$SUPPORTING_EVI@micro+=""
$CONTRADICTING_EVI+=""
$CONTRADICTING_EVI@cyto+=""
$CONTRADICTING_EVI@mito+=""
$CONTRADICTING_EVI@micro+=""
$EC=""
$GENES+=""
$TRANSCRIPTS+=""
$PROTEINS+=""
$REQUIRES+=""
$TODO+=""
$NOTES+=""
Now choose a Pubmed reference and a quoted statement from the abstract and insert it into
$SUPPORTING_EVI+="...".
Add more lines by adding more $SUPPORTING_EVI+="...".
Finally add a compartment specific reference. You can now observe the attributes assigned to the biochemical reaction by
clicking: "Menu-bar>Datasets>Focused_Dataset>Reactions defined by this dataset".
The following shows an example of a form for hexokinase filled out by the curator
Some text
$SUPPORTING_EVI+="PMID8717435 shown hexokinase in human liver ESCORE=5"
$SUPPORTING_EVI@cyto+="PMID7150652 cytosolic activity ESCORE=5 "
$SUPPORTING_EVI@mito+="PMID6035519 Appreciable activities were observed in mito... ESCORE=0 "
Searching PUBMED{ hexokinase AND mitochon* } yields the following:
$CONTRADICTING_EVI@mito+="Attached to outer mito membrane. Not in matrix. LOC{ ENSG00000106633 } ESCORE=3"
$EC="2.7.1.1 2.7.1.2"
$GENES+=" ENSG00000106633 ENSG00000156510 ENSG00000159399 ESCORE=5"
$TRANSCRIPTS+="ENST00000292432 ESCORE=5"
$PROTEINS+=" ALIGN{ UNIPROT:HXK1_HUMAN UNIPROT:HXK2_HUMAN UNIPROT:HXK3_HUMAN UNIPROT:HXK4_HUMAN } ESCORE=5"
$TODO+="Andreas: Please create a datasets for fructose. "
$NOTES+="NTPs other than ATP possible"