Assemble2 Features
-
A Complete 3D Modeling Workflow
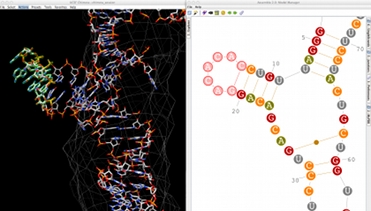
Assemble 2 allows you to construct your RNA 3D model interactively, starting from a solved tertiary structure and the orthologous sequence to model. But you can also use Assemble2 for more simple tasks: the prediction and manipulation of a secondary structure, the annotation and exploration of a solved tertiary structure.
-
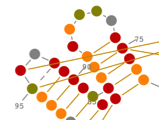
Prediction and Edition of 2D Structures
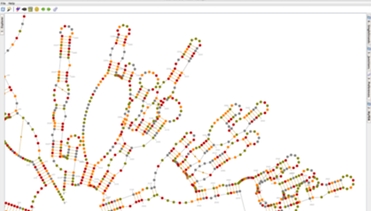
In Assemble2, your RNA 3D model is based on a secondary structure that can be loaded from a file or predicted using RNA algorithms wrapped as Web Services. Once loaded into Assemble2, this secondary structure can be edited interactively or used to explore a solved tertiary structure.
-
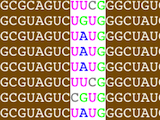
Construction of Structural Alignments
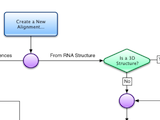
Assemble2 provides the alignment panel that was initally available with S2S. You can align an orthologous sequence against a solved tertiary structure, infer automatically its 2D and 3D structures, and pursue the modeling process without leaving Assemble2.
-
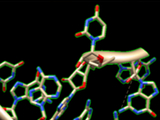
Guided 3D Modeling
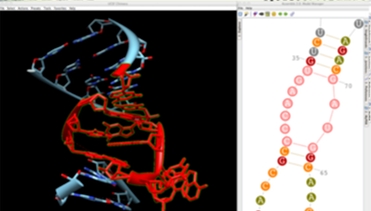
The RNA secondary structure allows you to define and select modules (helices and junctions) and to create their 3D fold directly in Chimera. For RNA junctions like apical loops, you can easily switch between different 3D folds extracted from solved tertiary structures and choose the one that fits best your assumptions. Chimera assembles a new 3D module automatically if a perfect match of module ends is found in the 3D model.
-
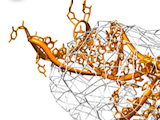
3D Modeling in Density Maps
Using the connection between Assemble2 and Chimera, you can use the fantastic vizualisation abilities of Chimera to guide the construction of your 3D model. Load a density map into Chimera, select an RNA junction into Assemble2 and switch between the different 3D folds to find the one that fit best your experimental data.
-
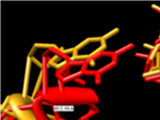
Built-in 3D Refinement
Assemble2 provides a geometric refinement function to optimize structural parameters like nucleotide stereochemistry, sugar pucker, and atoms distances deduced from the base–base interactions defined in the 2D panel. You can refine from few residues to the full 3D model. The refined structure is added to Chimera as a new layer, allowing you to compare 3D folds before and after refinement.
-

Powered by Web Services
Assemble2 is a semi-automatic environment where several tasks (RNA 2D prediction, RNA 3D modules search,...) are delegated to Web Services. You can use the public Web Services or deploy your own ones with the RNA Science Toolbox project. .
-

Run Everywhere
Assemble2 has been developed with the Java language on MacOSX. But it can run on any operating system having a Java environment.