Assemble2 Manual
Assemble2 is a Java application allowing you to compute, display, manipulate and edit RNA secondary structures interactively. Since it is linked to the 3D viewer UCSF Chimera, the secondary structure can be used to navigate into solved RNA tertiary structures or model/construct new ones.
Assemble2 is a semi-automatic environment in the sense that several tasks realized in Assemble2 are delegated to Web services hosted on our server:
- 2D prediction,
- 3D annotation,
- search for 3D fragments extracted from solved 3D structures,...
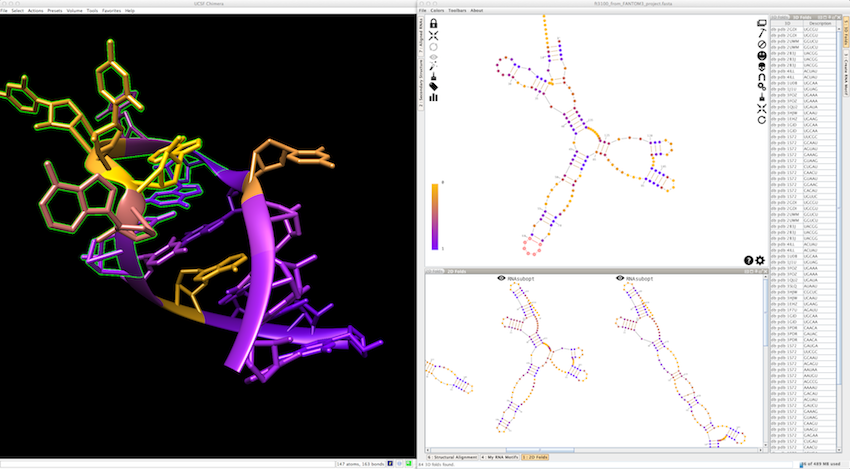
In the screenshot below, you can see that Assemble2 is made with several panels, several toolbars with icons and several menus. The 3D viewer UCSF Chimera (displayed on the left) is connected to Assemble2. Consequently, any selection done in the secondary structure displayed in the main panel highlights its 3D counterpart in Chimera.
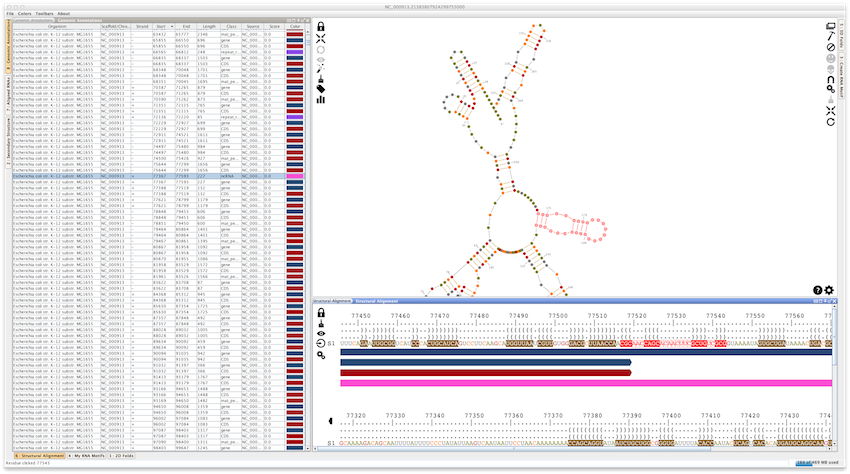
With its ability to load Genbank and GFF3 files, Assemble2 can also be used as a genome browser, allowing you to visualize and predict RNA structures for genomic annotations and/or selections. In the screenshot below, the complete genome of Escherichia coli str. K-12 substr. MG1655 has been loaded directly from the NCBI (File -> Load... -> Genomic Annotations -> From the Genbank Database).
In this manual, you will find more details about:
You will also find several tutorials:- how to install Assemble2?
- how to load data into Assemble2?
- how to manipulate an RNA secondary structure?
- how to edit an RNA secondary structure?
- how to derive a new 3D structure from a solved 3D? (modeling by homology)
- how to construct a 3D model from scratch? (ab initio modeling)
- how to refine a 3D model?
- how to construct and manipulate a structural alignment?