Tutorials
How to derive a new 3D structure from a solved 3D?
You have: a solved tertiary structure.
You want: to thread a new molecule into this tertiary structure (a.k.a. molecular replacement).
Depending on how close are the molecule extracted from the tertiary structure and the molecule you want to thread in (the target molecule), you have two options:
- if you just need to do subtle modifications, you can duplicate the reference molecule, do the modifications and thread it into the 3D structure,
- otherwise you need to add a new RNA molecule to the alignment, align it against the reference molecule and thread it into the 3D structure.
Duplication of the reference molecule
As an example, we will add a residue and mutate a second one in the D-loop of a tRNA. First, you need to load the file 1ehz.pdb provided with Assemble2 and available in "samples/tertiary structures". In the screenshot below, you can see the selection of this D-loop.
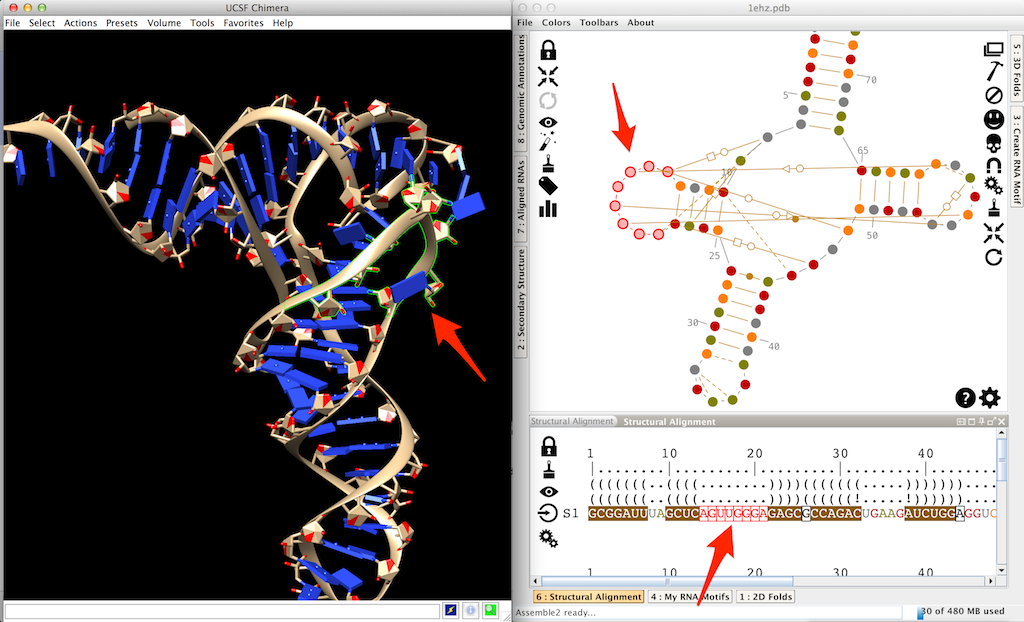
Open the lateral panel "Aligned RNAs", right-click on the reference molecule and choose "Duplicate".
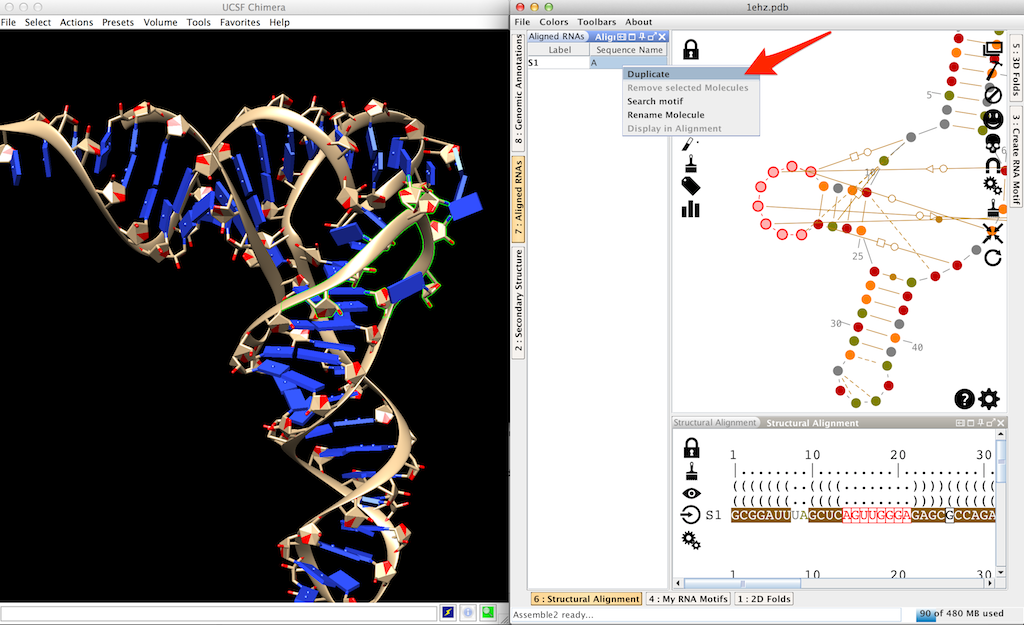
You can see that the structural alignment contains now a second molecule (labeled 'S2') whose sequence is identical to the first one.
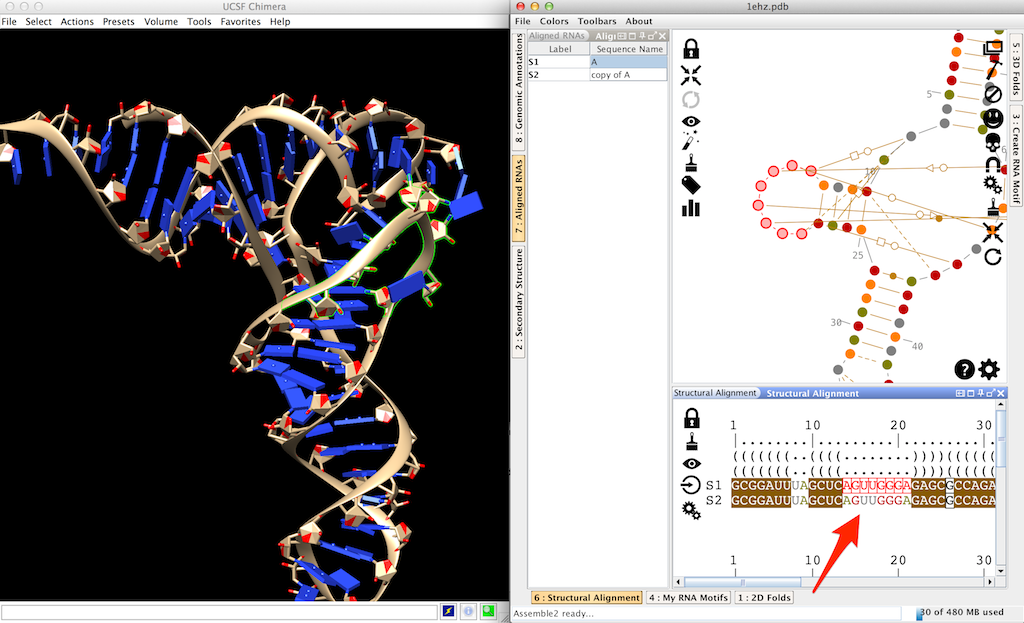
Click on to unlock the structural alignment.
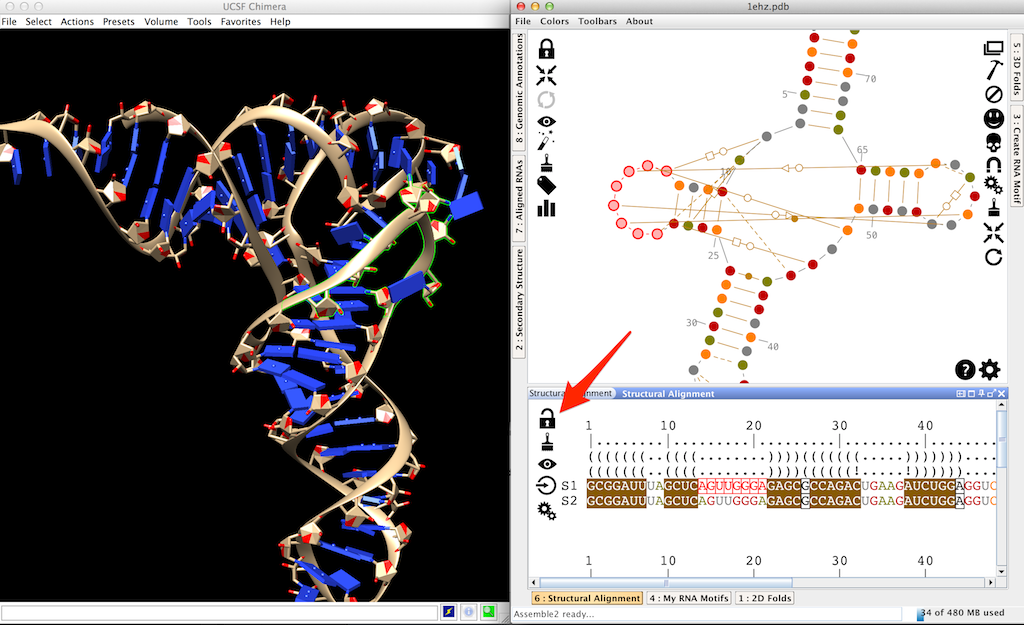
Click on the top row of the structural alignment (the row describing the positions of the alignment) to insert a new column.
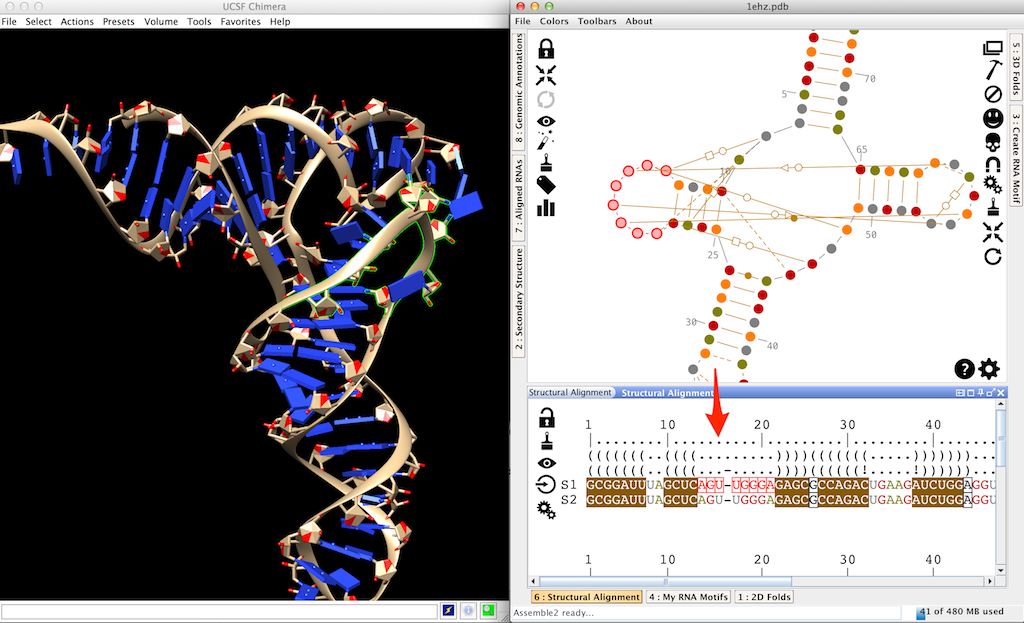
Click on the gap in the sequence 'S2' until you get a 'U'. This is an insertion.
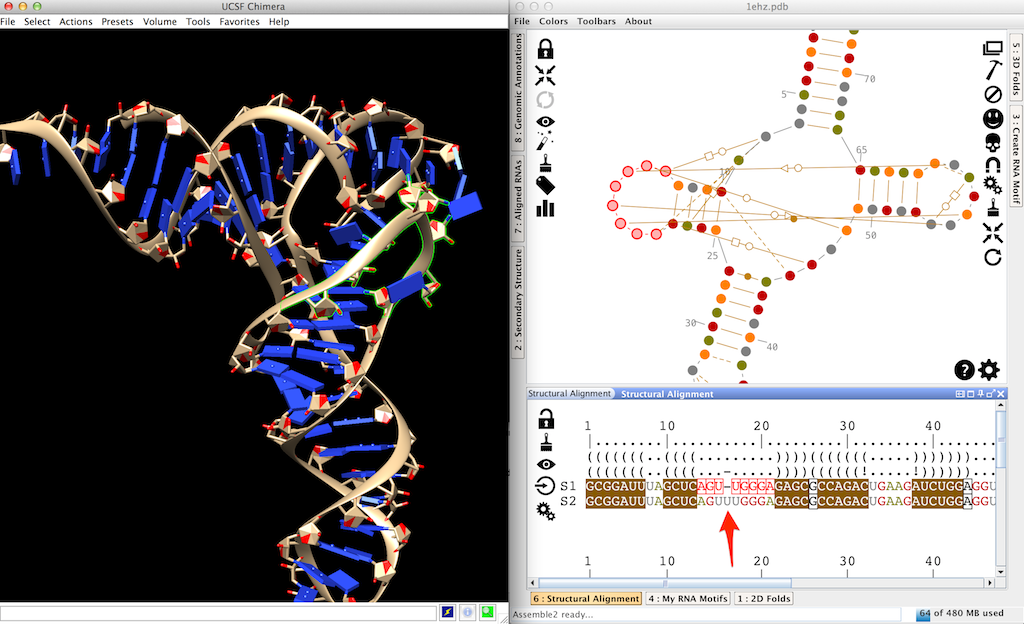
Click on the upstream 'G' in the sequence 'S2' until you get a 'A'. This is a mutation.
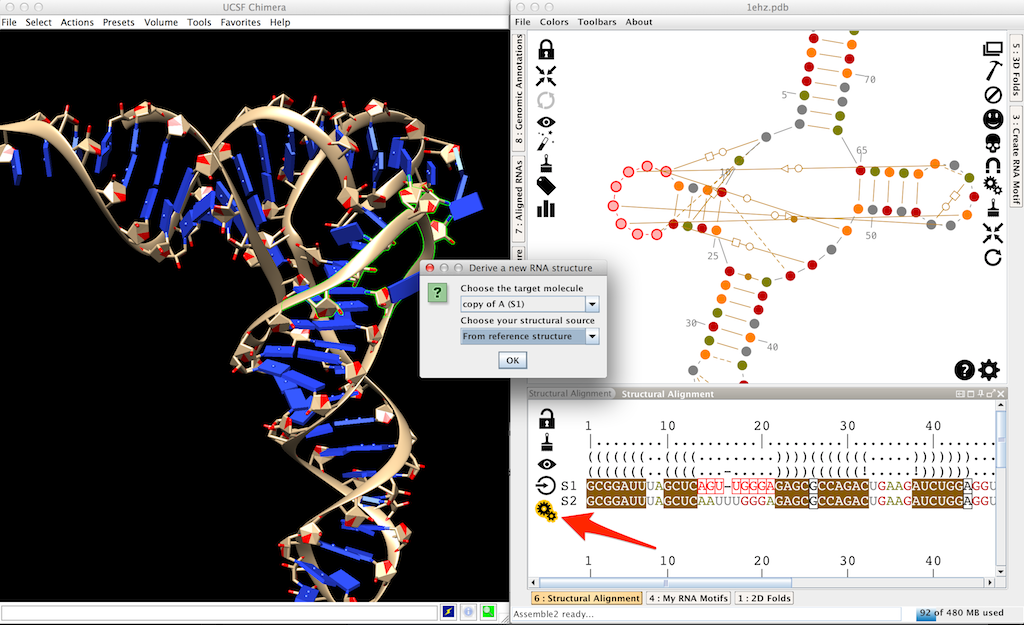
Click on to infer the new structure. This will popup a window. Choose 'S2' as the target molecule (the molecule you want to thread in the reference structure) and choose the reference structure.
Once the computation done, you can see that your target molecule is now the reference one (labeled 'S1'). The 3D and 2D didn't change much, but in the 2D you can see that the D-loop contains now an additional residue and a mutated residue. During the threading, the mutated residue has been processed. But the additional residue is still missing in the 3D structure.
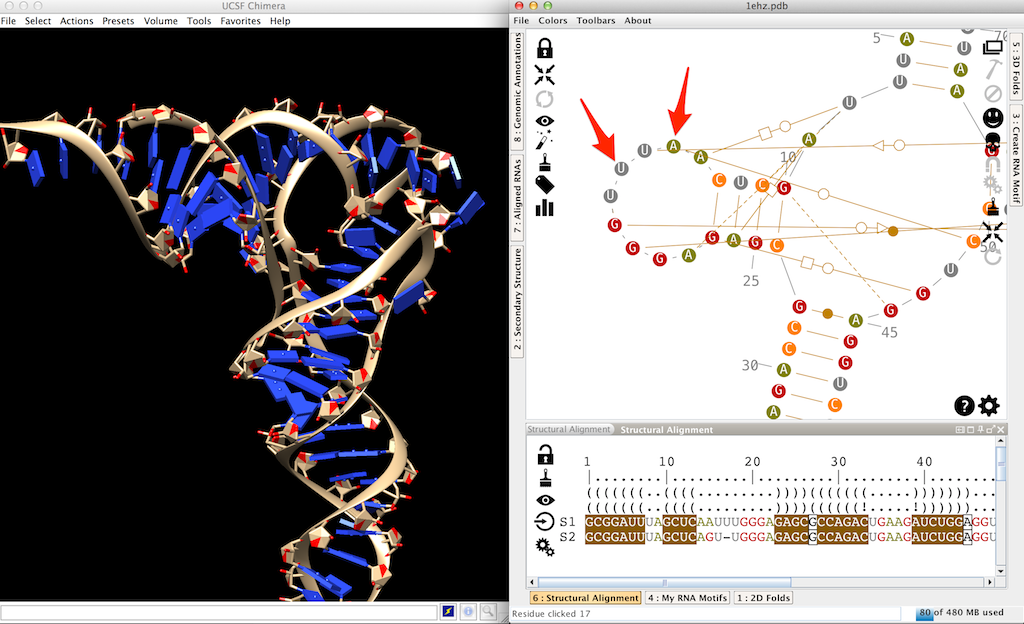
Select this residue in the main panel or in the lateral panel "Structural alignment" (arrow 1 in the screenshot). Click on to export this residue in Chimera (arrow 2 in the screenshot). Since the upstream residue is already present in the 3D structure, the new residue is stacked automatically (arrow 3 in the screenshot).
Instead to improve the placement of this residue by ourselves, we can use the ability of Assemble2 to refine the structure. See the tutorial "how to refine a 3D model?" to see how we will finalize this 3D structure.
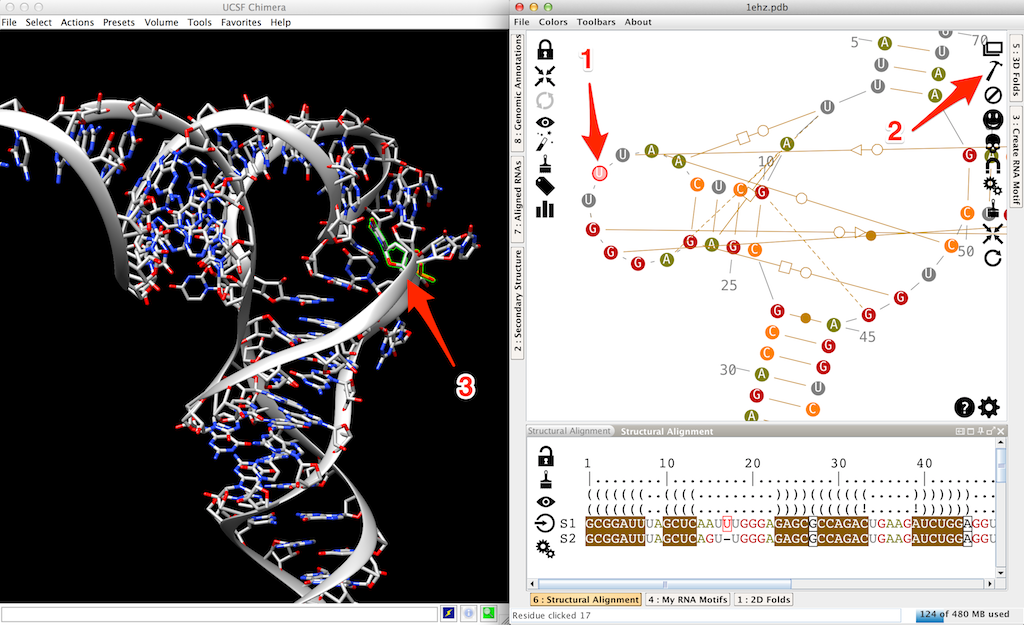
Another option would be to search for an apical loop of 9 nucleotides, extracted from a solved 3D structure, and to apply it to our mutated tRNA. To do so, we need first to erase the 3D residues for this loop. You can for example left-click on the first one and [shift]left-click on the last one (arrow 1 in the next screenshot). Then click on (arrow 2 in the screentshot) to trash these residues (arrow 3 in the screentshot).
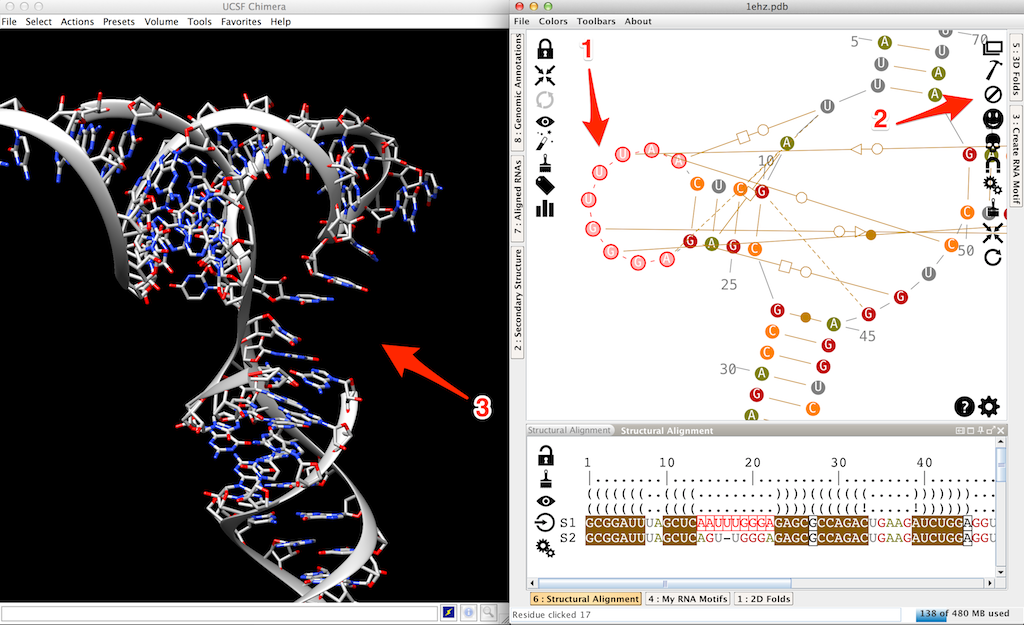
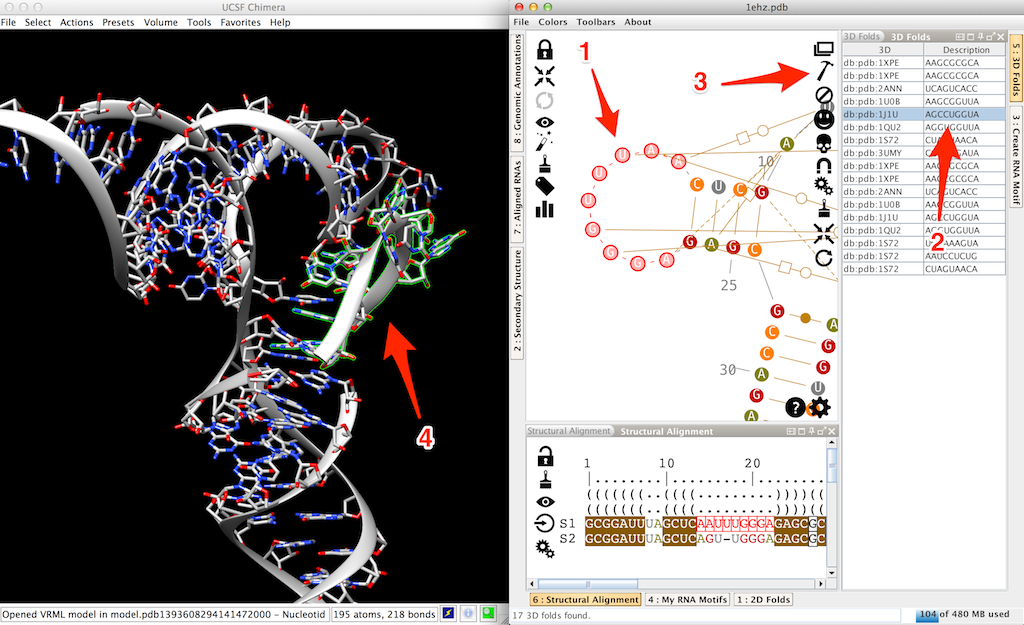
Now click twice on a residue of the D-loop (arrow 1 in the next screenshot). This will pop up the lateral panel "3D folds". Select an entry (arrow 2 in the screenshot) and click on (arrow 3 in the screenshot). Since the "sticky" residues are already present in the 3D (residues 13 and 23 in this example), the 3D fragment is automatically stacked on the helix (arrow 4 in the screenshot).
Alignment of a new RNA molecule
You have to follow these steps:
- add a new molecule in the alignment by clicking on
in the toolbar "Structural Alignment"
- align your molecule by moving the residues and/or adding gaps
- as described above, thread your RNA molecule in the 3D structure by clicking on
in the toolbar "Structural Alignment"
