Tutorials
How to construct and manipulate a structural alignment?
You have: a structural alignment.
You want: to play with it!!
In this tutorial, you will learn how to:
Once a structural alignment loaded (for more details, see the tutorial "how to load data into Assemble2?"), it is displayed in the lateral panel named "Structural Alignment". It displays all the sequences aligned against the reference sequence (and labeled 'S1'), which is folded and displayed in the main panel.
This panel allows you to contruct/improve a structural alignment interactively for a set of orthologous RNA sequences. Using colored structural masks, it will help you to identify base-pair covariations, leading to the characterization of a consensus secondary structure.
If the reference sequence ('S1') is linked to a tertiary structure, this panel allows you to derive it into a new RNA 3D model for one of the sequences aligned (for more details, see the tutorial "How to derive a new 3D structure from a solved 3D?").
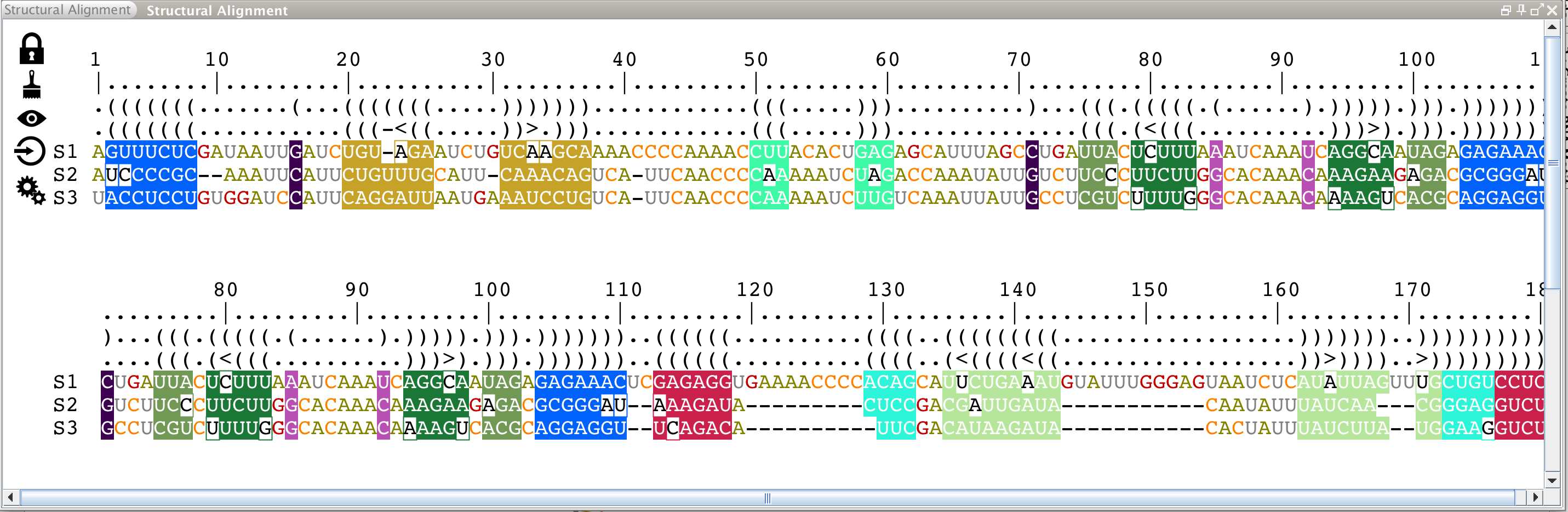
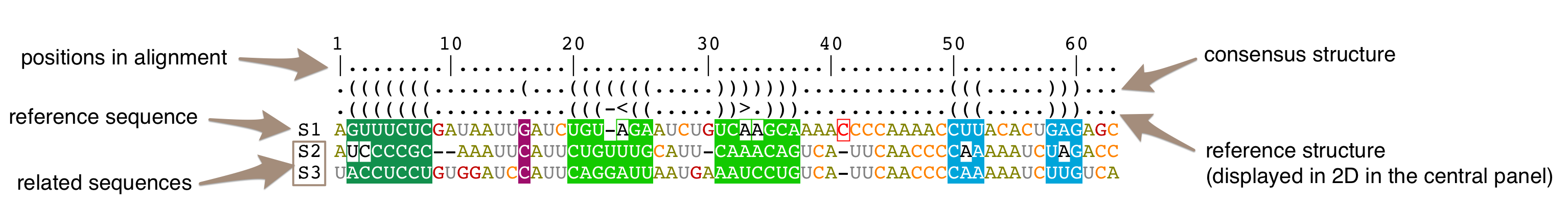
From top to bottom, a structural alignment is made with:
- a ruler for the alignment,
- a bracket notation describing the consensus secondary structure for all the sequences in the alignment,
- a bracket notation describing the secondary structure for the reference sequence (labeled as 'S1'). This bracket notation is a 1D notation of the secondary structure displayed in the main panel,
- the reference sequence for the alignment (labeled as 'S1'). It corresponds to the sequence you have selected when you have loaded your data. You can choose a different reference sequence with the icon
in the Alignment toolbar
- all the other sequences.
The consensus bracket notation is editable and describes:
- unpaired residues with the character '.'
- paired residues with the characters '(' and ')'
- unpaired residues with the character '.'
- paired residues in helices with the characters '(', ')', '[', ']', '{' and '}' (as in the lateral panel "Secondary Structure")
Manipulate a structural alignment
You can interact with your structural alignment using:
- the mouse and keyboard
- the alignment toolbar
Browse the alignment
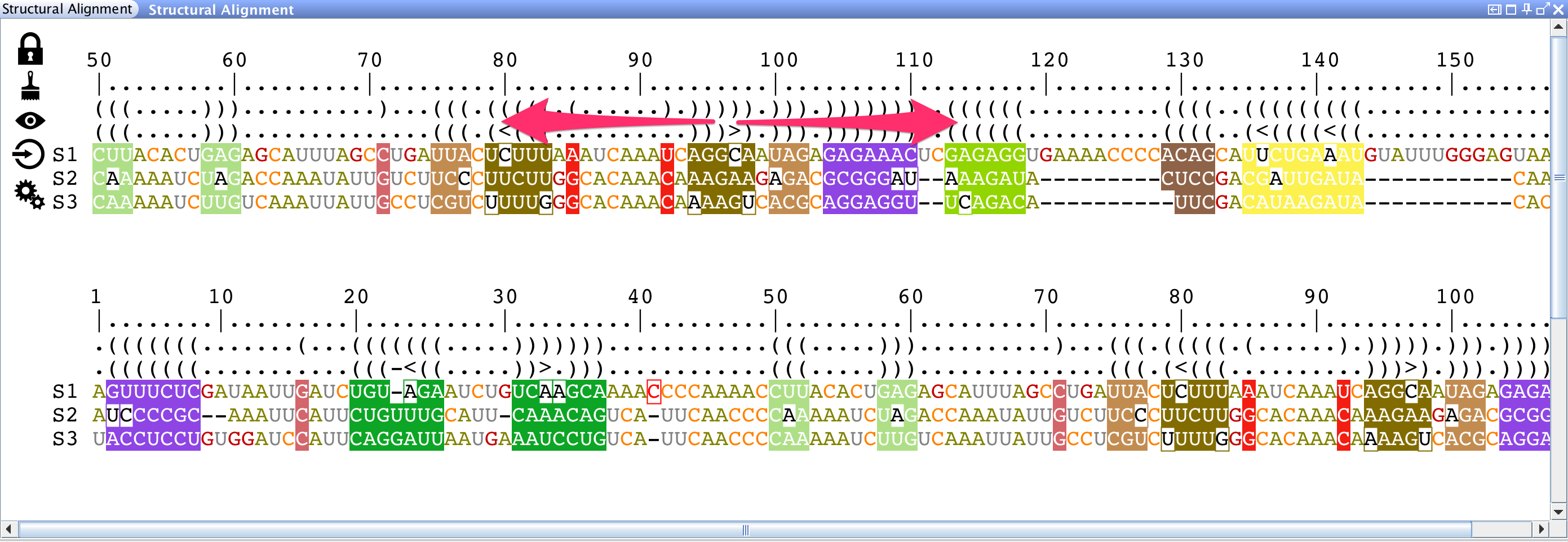
The lateral panel "Structural Alignment" splits the alignment into two views that can be manipulated independently. You can:


Or you can:

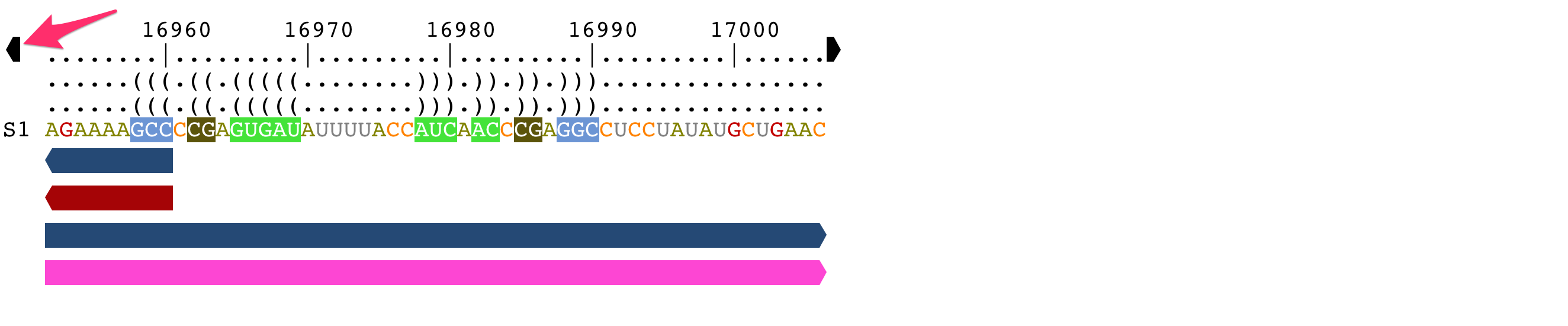
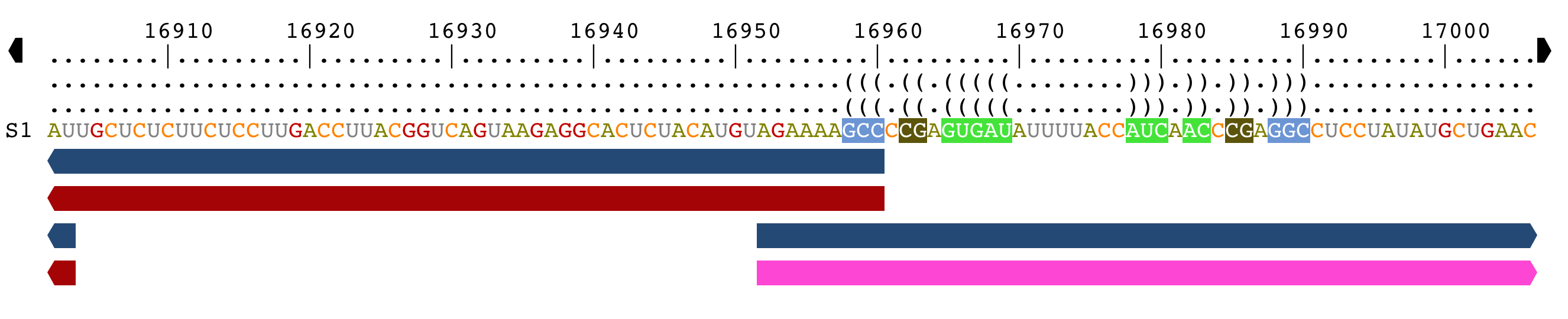
Browse genomic sequences
If you have opened a GFF3 or a Genbank file, and loaded one of the genomic annotation, two arrows are displayed at each end of the alignment.
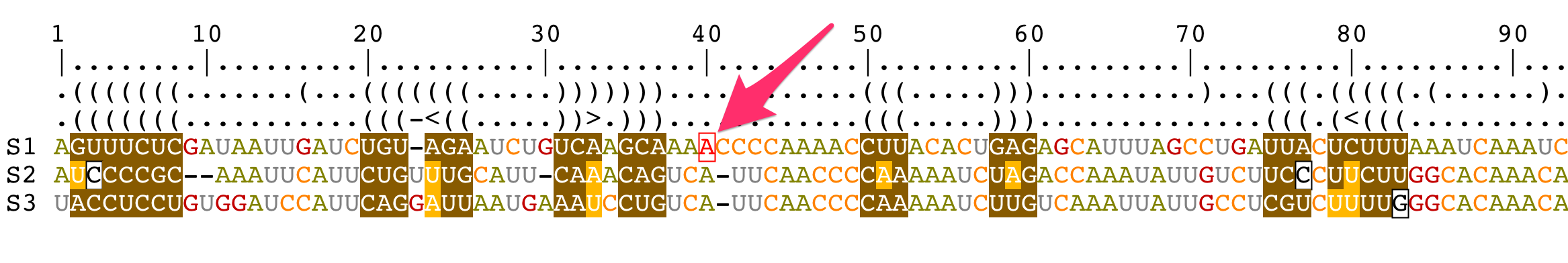
Select residues
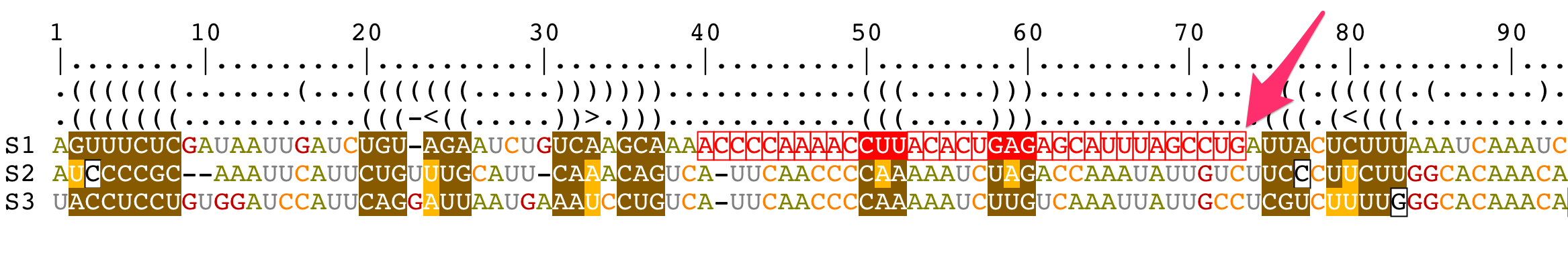
Modify a structural alignment
Move residues
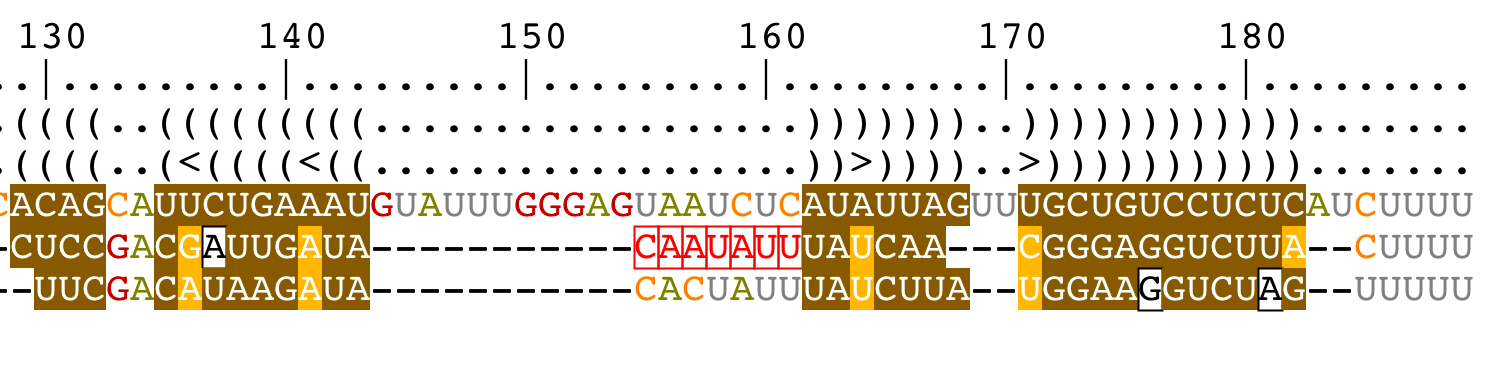
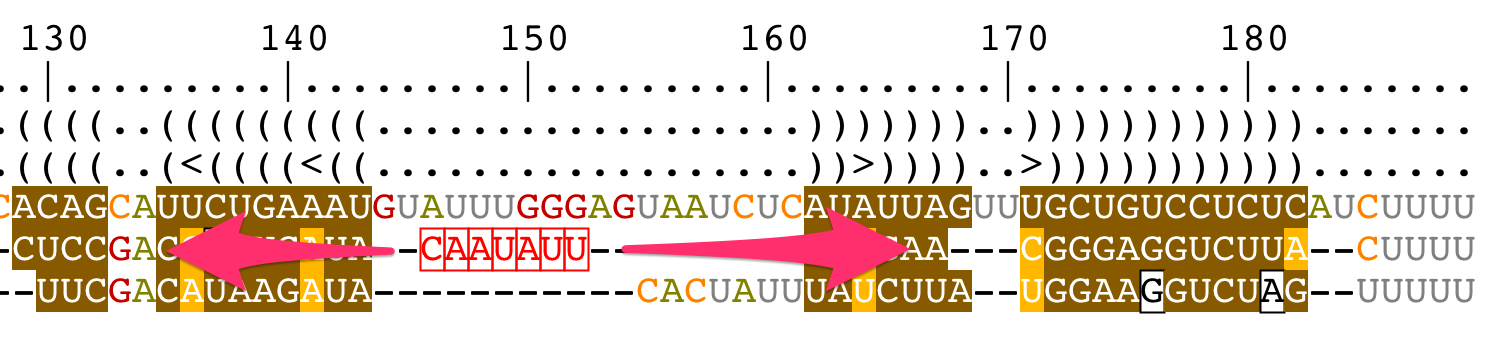
Add/remove gaps
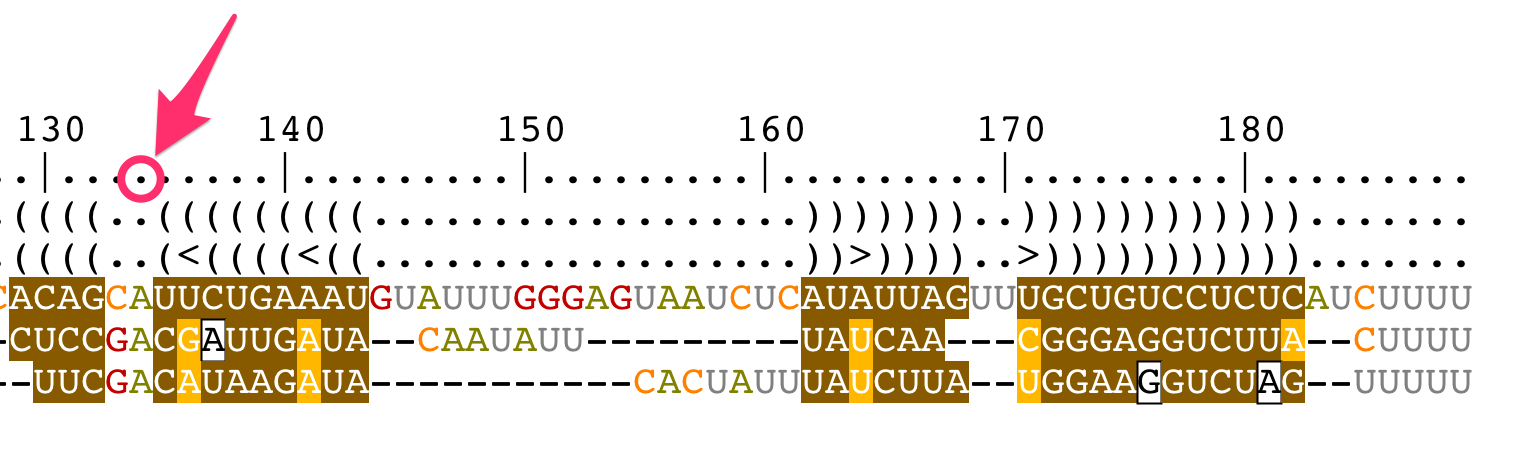
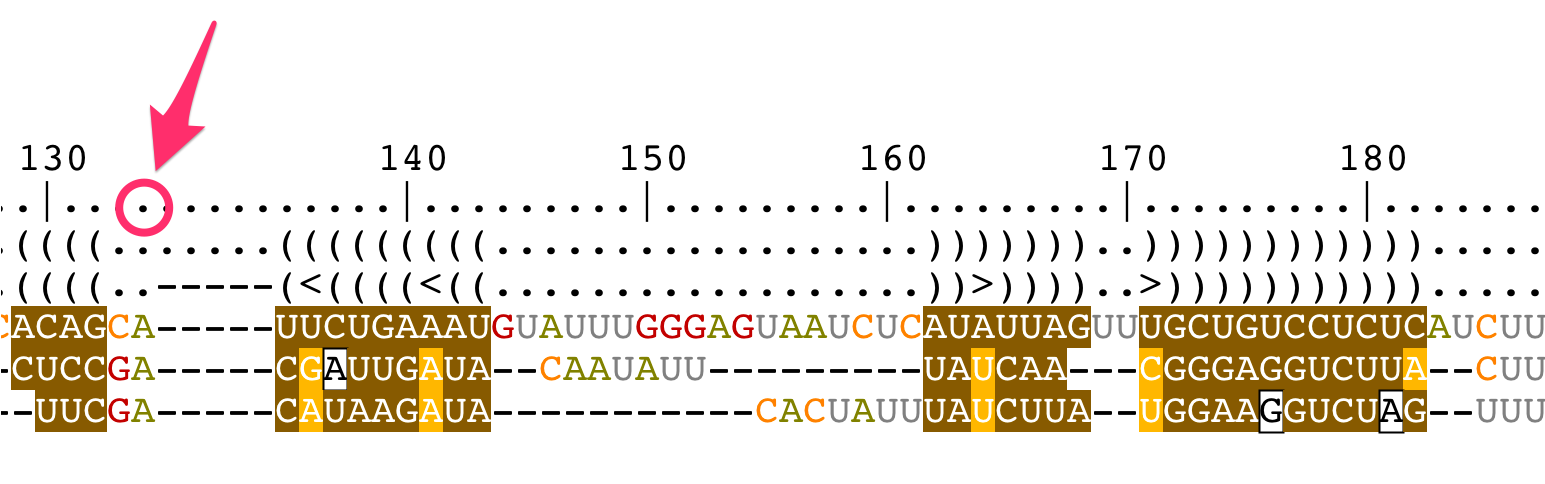

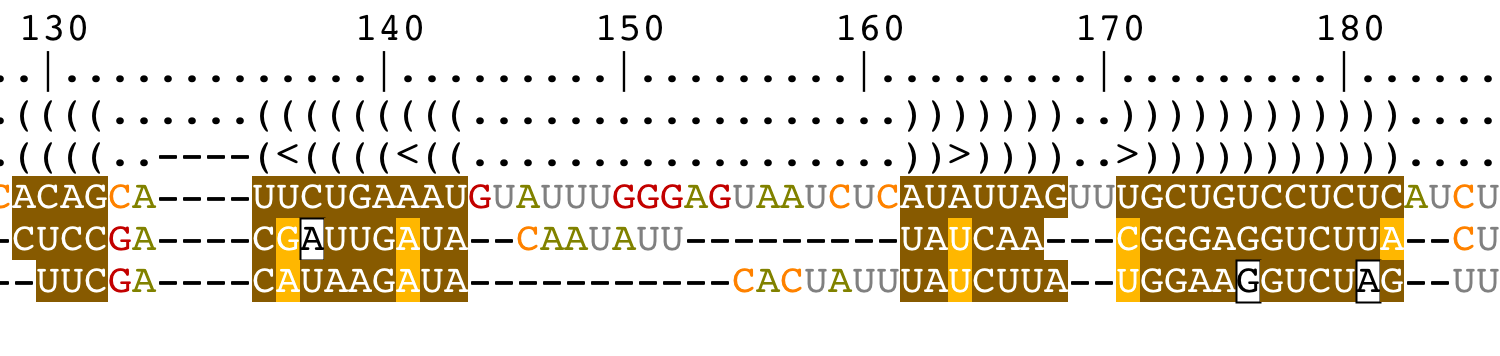
Change the bracket notations
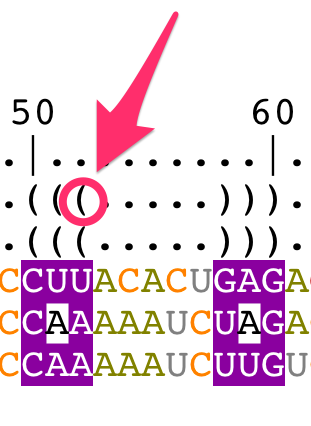
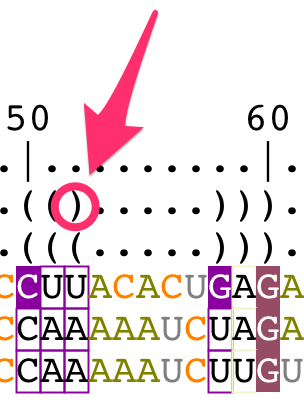
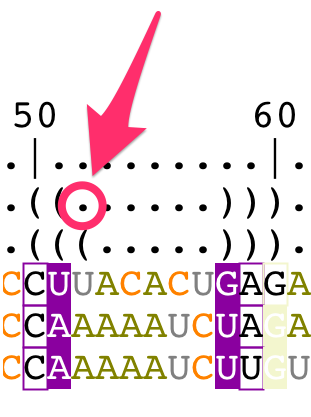
The bracket notation for the reference structure cannot be modified from the structural alignment. You have to edit the secondary structure directly from the main panel (for details, see the tutorial "How to edit an RNA secondary structure?").

