The Panels
You will find details about the panels of Assemble2:
The main panel
Once a secondary structure loaded (for more details, see the tutorial "how to load data into Assemble2?"), it is displayed in the main panel.
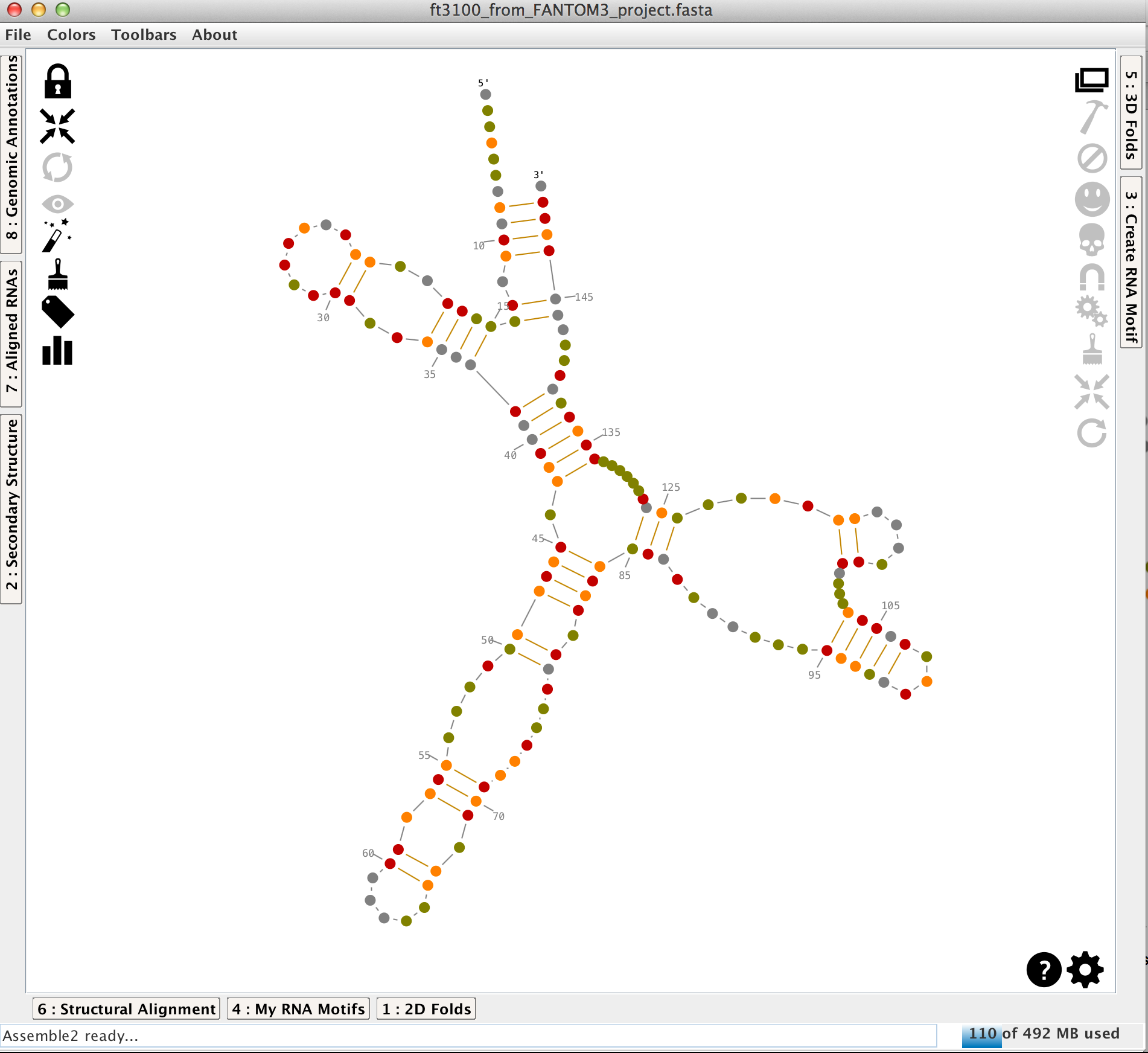
A secondary structure is made with helices linked and oriented by single-strands and junctions (apical loops, inner loops, 3-way junctions,...). In Assemble2, the helices can be moved to modify the layout of the secondary structure.
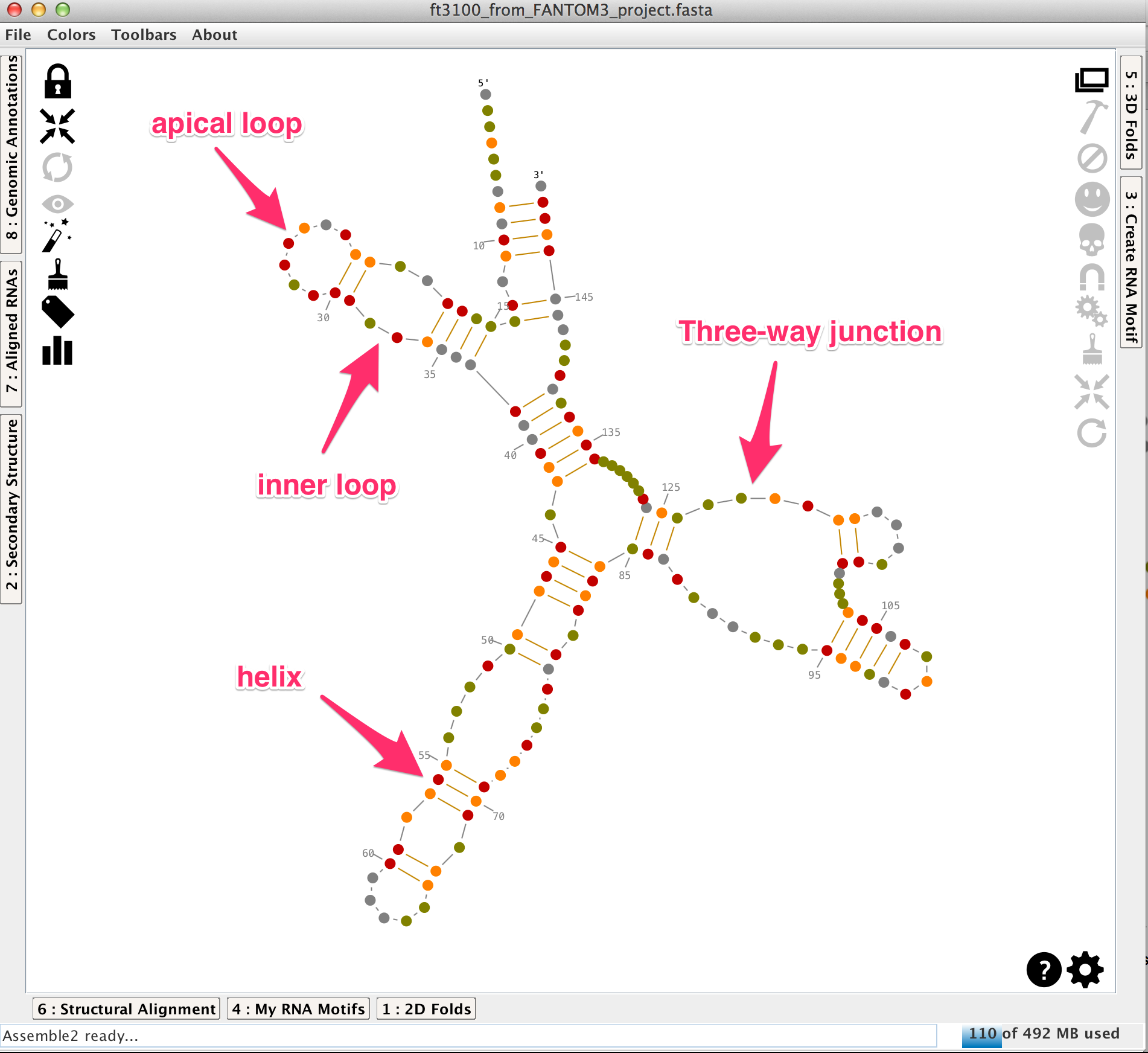
The main panel displays also the 2D toolbar (on the left) and the 3D toolbar (on the right). You can interact with the secondary structure using the 2D toolbar or the mouse. The 3D toolbar is used to export and manipulate pieces of the secondary structure as 3D folds into Chimera.
To learn more about the manipulation of an RNA secondary structure, take a look at the tutorial "how to manipulate an RNA secondary structure?".
The lateral panels
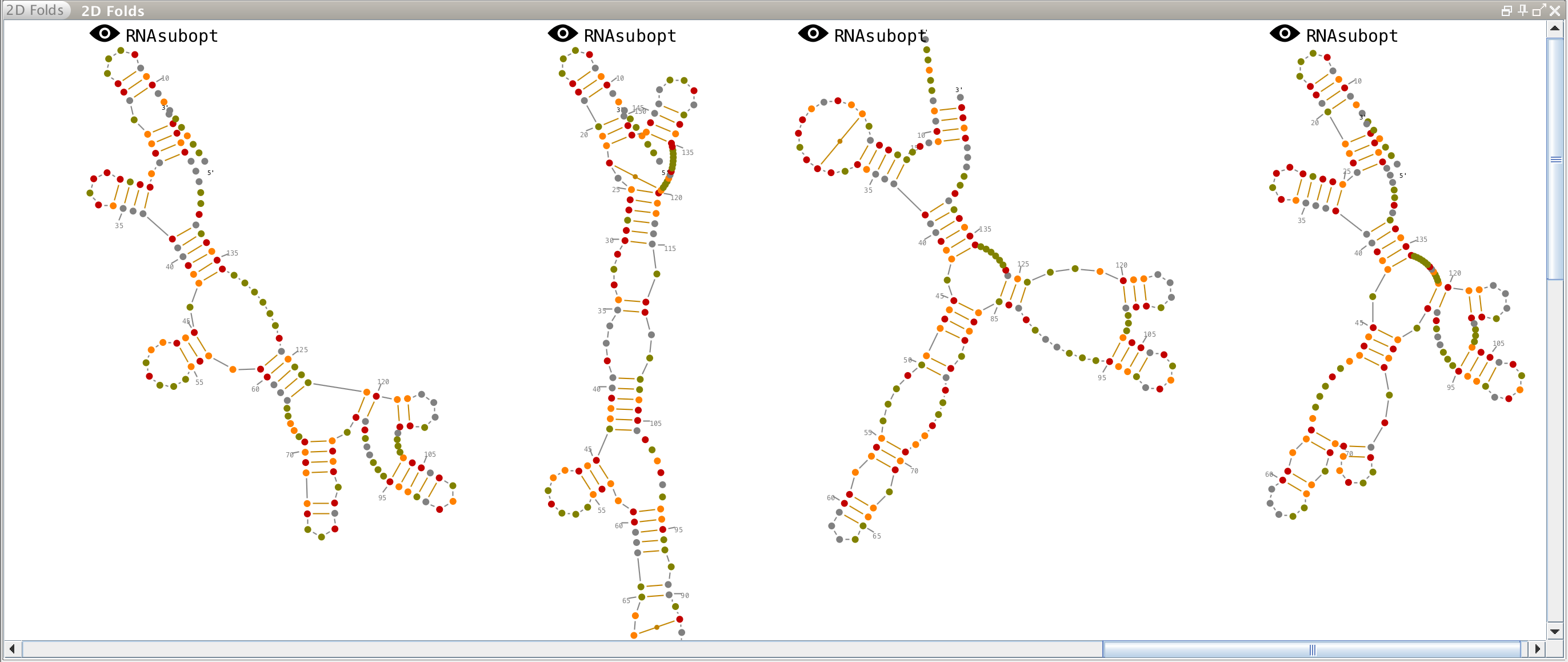
2D folds
This lateral panel is displayed automatically when a secondary structure is predicted from one or several molecules (see the tutorial "How to load data into Assemble2?").
Each prediction is linked to this icon and along with the name of the algorithm that produced this prediction. To choose a secondary structure to work with, you just have to click on
.
Structural Alignment
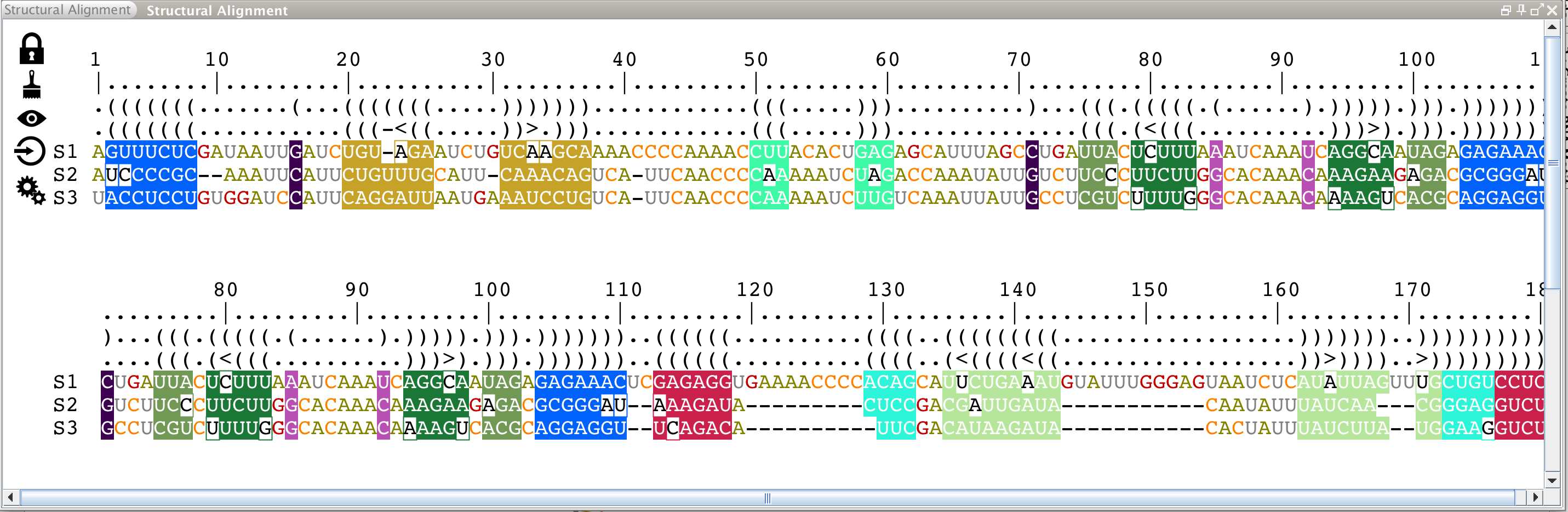
This panel displays all the sequences aligned against the reference sequence (and labeled 'S1'), which is folded and displayed in the main panel. These sequences can have been loaded directly from an alignment file (Clustal file), from a database entry (RFAM database) or imported afterwards from the lateral panels named "Aligned RNAs" (see next paragraph).
To learn more about the manipulation of a structural alignment, take a look at the tutorial "how to construct and manipulate a structural alignment?".
Aligned RNAs
This lateral panel list all the sequences available in the structural alignment. By right-clicking on one of these sequences, you can:
- Duplicate: the reference molecule (labeled 'S1') can be duplicated, allowing you to derive its structure. For more details, see the tutorial "How to derive a new 3D structure from a solved 3D?",
- Remove Selected Molecules: will remove, from the structural alignment, all the molecules selected in this panel (except the reference molecule labeled 'S1'),
- Search Motif: a window will popup, allowing you to describe a sequential motif to be searched in all the molecules selected in the list. You can use regular expressions to describe your motif. If matches are found in the reference molecule ('S1'), they will also be highlighted in the secondary structure displayed in the main panel, and in the 3D structure displayed in Chimera (if any).
- Rename Molecule: should be explicit enough ;-)
- Display in Alignment: the two views in the panel "Structural Alignment" will update to display all the molecules from the first one to this one.
Secondary Structure
This lateral panel lists hierarchically all the elements making the secondary structure in the main panel. By left-clicking on one of these elements, it will be selected in the main panel. By right-clicking on one of these elements, you will get the following choices:
- Rename: this allows you to rename molecules, helices and single-strands,
- Set color: define a new color for the element,
- Clear color: the element will recover its default display
- '(' and ')' for Watson-Crick edges,
- '[' and ']' for Hoogsteen edges,
- '{' and '}' for Sugar edges.
3D folds
This lateral panel is displayed automatically when a single-strand or a junction is selected (see the tutorial "how to manipulate an RNA secondary structure?" for details about selections). Once the selection done, Assemble2 queries a database hosted on our server for 3D fragments extracted from the Protein DataBank and compatible with the single-strand or junction selected.
Create RNA Motif
If you need to apply a specific 3D fragment in your 3D model, you can extract and store it yourself in a local repository.
My RNA Motifs
This lateral panel allows you to browse and manage your local repository of 3D fragments.
Genomic Annotations
If you right-click on a genomic annotation, Assemble2 will compute its folding landscape and display it in the lateral panel "2D folds". Be careful of the size of the genomic annotation, otherwise the computation could take a while.
Chimera
The 3D viewer UCSF Chimera is not exactly a panel like the previous ones. It is a software you have to install on your computer and to connect to Assemble2. First you need to record the absolute path for the Chimera executable:
- either at the first launch of Assemble2 (a window will popup to set the path for the executable of Chimera),
- or from the menu "File -> Configure... -> Assemble2" (see here for details).
- when you start Assemble2
- by clicking on
(see here for details)